Exploring the Therapeutic Potential of Metal Complexes of Pregabalin and Terbutaline: Spectroscopic Insights and Molecular Docking in Alzheimer’s and Parkinson’s Disease
Parth Anilkumar Barot* , Mohyuddin Abdulbhai Maradiya
, Mohyuddin Abdulbhai Maradiya and Jabali J. Vora
and Jabali J. Vora
Department of Chemistry, Hemchandracharya North Gujarat University, City-Patan, State-Gujarat, Country- India.
Corresponding Author E-mail: barotparth3333@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/400219
Article Received on : 10 Feb 2024
Article Accepted on :
Article Published : 29 Mar 2024
Reviewed by: Dr. Achal Mishra
Second Review by: Dr. Kanagathara Narayanan
Final Approval by: Dr. Dinesh Chand Sharma
The study investigated the formation of cobalt, nickel, and copper metal complexes by reacting metal chloride with the ligand pregabalin and the medication terbutaline. The complexes had the formula C20H40N2O7MCl2, where M = Co(II), Ni(II), and Cu(II). The analysis used elemental data, molar mass, and infrared (IR) spectrum studies. The IR frequencies of the ligand bands showed changes, indicating coordination with the metal ion. The study found that the ligands pregabalin and terbutaline exhibited bidentate properties of metal complexes. The research focused on how metal complexation affected the functionality of terbutaline and pregabalin complexes about Alzheimer's Disease (AD) and Parkinson's Disease (PD). The results demonstrated strong interactions with amino acids in the binding region of the targeted protein. The study suggested that zinc metal carboxylates with antioxidant and anticholinesterase properties could be beneficial in treating Alzheimer's diseases.
KEYWORDS:Alzheimer’s disease; Metal complexes; Molecular Docking; Pregabalin; Parkinson’s disease; Terbutaline
Download this article as:| Copy the following to cite this article: Barot P. A, Maradiya M. A, Vora J. J. Exploring the Therapeutic Potential of Metal Complexes of Pregabalin and Terbutaline: Spectroscopic Insights and Molecular Docking in Alzheimer’s and Parkinson’s Disease. Orient J Chem 2024;40(2). |
| Copy the following to cite this URL: Barot P. A, Maradiya M. A, Vora J. J. Exploring the Therapeutic Potential of Metal Complexes of Pregabalin and Terbutaline: Spectroscopic Insights and Molecular Docking in Alzheimer’s and Parkinson’s Disease. Orient J Chem 2024;40(2). Available from: https://bit.ly/3TAdrYl |
Introduction
Pregabalin (PGB) is a neurotransmitter that is structurally related to gamma-aminobutyric acid and has chemical similarities with gabapentin (GABA). PGB is a gamma-amino acid and its 3-substituted derivative, (S)-(þ)-3-isobutyl-GABA, behaves similarly to GABA. Similar to the alpha-amino acids L-leucine and L-isoleucine, PGB has demonstrated effectiveness in treating various conditions, such as severe diabetic peripheral neuropathy and neuropathic pain. Understanding its pharmacodynamics may depend more on these a-amino acid similarities than on its structural resemblance to GABA 1, 2.
Terbutaline (TRB), a significant phenylethanolamine drug, is frequently used to treat allergic airway diseases, including bronchial asthma, both abruptly and over time 3-5. Because of its rapid body distribution, it causes bronchodilation, vasodilatation, and an increase in heart rate 6–8. However, researchers have also become interested in the negative consequences of TRB on respiratory ailments and other areas. Overdosing on TRB can result in fatal outcomes in certain cases, as well as headaches, shaking, and a rapid heartbeat 9,10. Another prohibited application of the medication is regulating its use to improve athletes’ anabolic effects and performance during sports 11.
Alzheimer’s disease (AD), one of the most common neurodegenerative disorders, ranks as the primary cause of dementia among adults. The health care system is severely strained socioeconomically by AD because of its atypical behaviors, cognitive deficits, and challenges carrying out daily tasks. AD is considered an advanced neurological illness. Memory loss and learning difficulties are among the disease’s primary symptoms. As of yet, there is no effective treatment for AD. The amyloid beta-peptide is pivotal in the development of amyloid plaques detected in the brains of those with Alzheimer’s disease (PDB:1IYT). Enzymatic cleavage differentiates it from the amyloid precursor protein (APP). Aβ40 and Aβ42 are the most well studied Aβ peptide variations, though there are others as well. Aβ42 is especially noteworthy because of its tendency to generate neurotoxic plaques and solidify 12. The primary relationship of the protein alpha-synuclein (PDB: 1XQT) with Parkinson’s disease is the formation of aggregates known as Lewy bodies 13. Another prominent form of neurodegenerative disease is Parkinson’s disease (PD), which damages the brain’s nigrostriatal circuit due to the production of Lewy bodies as byproducts and the absence of dopamine neurotransmitters.
An essential characteristic of a metal is its capability to generate positively charged ions in an aqueous solution, enabling them to subsequently bind to negatively charged biological molecules 14. In contrast to 3d-transition metal ion complexes, complexes of 4d- or 5d-metal ions are less toxic and have a greater ability to pass through microorganisms’ cell membranes 15. In order to interact with the TRB and PGB as a ligand, we have chosen a set of two divalent metal ions, M(II), such as Cu2+, Ni2+, and Co2+, as the main metal ions in our work (L). To provide insight into the structures and bonds that were formed, the current study used FT-IR, elemental analysis, mass spectrometry, and molecular modelling of novel mixed ligand complexes.
Material and Method
Materials and Physical Technique
Since all of the chemical reagents used in the experiments performed were analytical grade, no additional purification was necessary. After a 24-hour soak in a chromic mixture (K2Cr2O7 + conc. H2SO4), glassware was carefully cleaned with double distilled water and dried at 120 °C in an oven. Terbutaline, Pregabalin, CoCl2.6H2O, NiCl2.6H2O, and CuCl2.2H2O, Methanol. Melting points, acquired via a DBK-programmed melting point apparatus, remain uncorrected. The synthesized compounds underwent purification through recrystallization using a suitable solvent system. Purity assessment employed the TLC technique, with spots visualized through exposure to iodine vapors and a UV cabinet.
Synthesis
A stirred suspension containing 1 mmol (0.16 g) of PGB and 1 mmol (0.37 g) of TB in 50 ml of methanol was gently supplemented dropwise with 1 mmol (0.26 g) of CoCl2H2O in 20 ml of methanol in order to synthesise the solid metal complex [Co(TRB)(PGB)(H2O)2]Cl2 (A). The precipitate was filtered, repeatedly washed with methanol, and vacuum-dried using anhydrous calcium chloride following a six-hour reflux of the reaction mixture.. Using CoCl2.6H2O, NiCl2.6H2O, and CuCl2.2H2O in methanol, respectively, in a 1:1:1 (M+2:TRB:PGB) molar ratio, the solid complexes [Co(TRB)(PGB)(H2O)2]Cl2 (A), [Ni(TRB)(PGB)(H2O)2]Cl2 (B), and [Cu(TRB)(PGB)(H2O)2]Cl2 (C) were produced in a manner similar to that previously described 16. Metal complexes’ molecular structure is depicted in Figure 1.
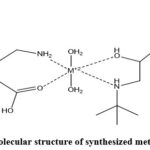 |
Figure 1: Molecular structure of synthesized metal complexes |
Characterization
Fourier Transform Infrared Spectroscopy (FT-IR)
Functional group elucidation was carried out employing a Fourier Transform Infrared spectrophotometer (FT-IR) model Shimadzu-8400. The FT-IR analysis was conducted utilizing an FTIR analyzer at Vaibhav Analytical Lab in Ahmedabad, with 100 scans executed within the spectrum range of 4000–400 cm-1.
Elemental Analysis
Precisely measure the sample weight using an analytical balance, ensuring that the sample size aligns with the sensitivity requirements of the elemental analysis technique. Introduce the accurately weighed sample into the combustion chamber of the Perkin-Elmer model 240.B. Employ a carefully controlled combustion process to transform the metal complex sample into gaseous products. Channel the resultant combustion gases through a separation column designed to isolate individual elemental components (C, H, N, O).
Mass Spectrometry
The acquisition of mass spectrometry data involved the utilization of Shimadzu QP-2010-Plus (E170Ev) mass spectrometer, which comes equipped with a highly efficient electrospray ionization (ESI) interface. The subsequent analysis of the obtained data was performed using sophisticated MassLynx 4.1 software. The implementation of tandem mass spectrometry (MS/MS) for detection further enhanced the precision of the analysis, with the instrument operating in the electrospray positive ionization mode.
Molecular Docking Methodology
Receptor Preparation
The RCSB Protein Data Bank served as the primary source for obtaining the structural information of metal complexes associated with proteins implicated in Alzheimer’s and Parkinson’s diseases, namely, 1IYT, 2BEG, 1XQ8, and 2KKW 17-20. To optimize the receptor for subsequent molecular docking, a comprehensive series of preparatory steps were meticulously executed:
PDB File Retrieval
The RCSB website facilitated the acquisition of the Protein Data Bank (PDB) file corresponding to the designated protein.
PyRx Software Implementation
PyRx, a software application operating on a Windows 10 platform, specifically version 0.8, was employed for handling the subsequent molecular docking procedures. The receptor structure, in its 64-bit format, was seamlessly imported into the PyRx environment.
Preprocessing Stages
A thorough preprocessing phase was initiated to refine the receptor structure for optimal docking simulations. Water molecules, co-crystallized ligands, and any heteroatoms deemed nonessential for binding interactions were systematically removed from the protein structure.
Structural Refinement
To further enhance the receptor structure, adjustments were made to optimize link lengths and angles. Hydrogen atoms were judiciously added to the structure, contributing to an improved and more accurate representation of the receptor for subsequent molecular docking analyses.
By meticulously executing these preparatory measures, the receptor was meticulously fine-tuned, ensuring its suitability for the ensuing molecular docking investigations. These steps not only involved the extraction of relevant structural information but also incorporated advanced structural refinement techniques to bolster the accuracy and reliability of the molecular docking simulations.
Ligand Preparation
The synthesis of ligands, constituting a repertoire of synthetic hybrid compounds featuring metal complexes, involved a systematic series of steps designed for precision and accuracy:
Chemical Structure Design: Leveraging the authorized Ultra 11.0 edition of ChemDraw, the chemical structures of ligands were meticulously drawn and refined. This process unfolded on a 64-bit Windows 7 system, ensuring optimal software compatibility 21.
Structural Validation and Transformation: To guarantee the correctness of bond connections and protonation states, the ligand structures underwent a crucial transformation into the PDBQT format. This conversion step was essential for the subsequent molecular docking analyses, promoting accuracy in ligand-receptor interactions.
Energy Minimization Procedure: The prepared ligand structures underwent a rigorous energy minimization process using (BIOVIA DS Visualizer 4.5) 22. This step involved the optimization of molecular geometry and energy distribution within the ligands, ensuring their stability and conformational accuracy for reliable engagement in molecular docking simulations.
By adhering to these meticulous procedures, the ligands were systematically designed, validated, and refined, culminating in a set of synthetic hybrid compounds with metal complexes optimized for subsequent molecular docking investigations. The utilization of advanced software tools and the incorporation of an energy minimization step collectively contributed to the precision and reliability of the ligand structures for computational analyses.
Molecular docking
The molecular docking procedure was executed employing the AutoDock Vina module within the PyRx software, adhering closely to the guidelines outlined in references23 and24. In line with the methodologies specified in reference22, conformational data analysis of the protein-ligand complexes was undertaken using the Biovia Discovery Studio. This comprehensive approach ensured a robust exploration of molecular interactions, aligning with established protocols for reliable and insightful computational investigations.
Result and Discussion
Fourier Transform Infrared Spectroscopy (FT-IR)
Figure 2 displays the FT-IR spectra for metal complexes, TRB, and PGB. PGB’s FT-IR spectrum revealed an unique band at 3500–3600 cm-1 and 3430 cm-1, respectively, that was identified as the -COOH and -NH stretching vibrations. Around 1646 cm-1, the bands were seen; -C=O stretching was attributed to them, and -C-O stretching was assigned to them between 1210 and 1320. The C-H stretching vibration may be the cause of the asymmetric band that was seen between 2960 and 2850 cm-1 25 The FT-IR spectrum of TRB showed peaks at 3500-3600 cm-1, 1260-1000 cm-1, 3300-3500 cm-1, 2805 cm-1, 1611 cm-1, 1332.6 cm-1, and 1121 cm-1, respectively, for O-H stretching for phenolic, C-O stretching for alcoholic group, N-H stretching for amine group, C-H stretching for alkane, C-N stretching for amine group, C=C stretching for aromatic ring, and 2° O-H stretching of alcoholic group26.
Furthermore, the bands corresponding to the ligand found at 1608, 3328-3342, and 1107 cm-1 in each complex’s spectra showed shifts to lower frequencies of roughly (35-40), (80-90), and (12-15) cm-1, respectively. These bands were the ν(alcoholic) O-H of TRB, the v(amine) of PGB and TRB, and the v(carbonyl) of carboxylic acid. These changes demonstrate that the ligand was coordinated with the metal ions through the alcoholic group’s nitrogen atoms and the carboxylic acid’s oxygen atom. At lower frequencies in the complexes, new bands were found and identified as belonging to the υ(M-N) and υ(M-O), respectively, and approximately (485-480) and (410-420). The FT-IR spectrum of TRB showed peaks at 3500-3600 cm-1, 1260-1000 cm-1, 3300-3500 cm-1, 2805 cm-1, 1611 cm-1, 1332.6 cm-1, and 1121 cm-1, respectively, for O-H stretching for phenolic, C-O stretching for alcoholic group, N-H stretching for amine group, C-H stretching for alkane, C-N stretching for amine group, C=C stretching for aromatic ring, and 2° O-H stretching of alcoholic group26,27. Furthermore, it was shown that ω (M-OH2) was connected to stretching vibrations in the 833–696 cm-1 range. This provides compelling evidence that water molecules are involved in the coordinating process28. Every one of these statistics is shown in Table 1.
 |
Figure 2: FT-IR spectrum of PGB, TRB, (A), (B), and (C) complexes |
Table 1: Wavenumbers are used to represent the FT-IR spectra of the free ligand and its metal complexes (cm-1).
|
|
ѵ C=O |
ѵ O-H |
ѵ C=O |
ѵ O-H |
ѵ N-H |
ѵ M-O |
ѵ M-H |
H2O |
|
PGB |
1646 |
3500-3600 |
1210-1320 |
—– |
3430 |
– |
– |
— |
|
TRB |
—- |
3500-3600 |
1000-1260 |
1121 |
3300-3500 |
– |
– |
— |
|
Co |
1608 |
3500-3600 |
1210-1320 |
1107 |
3328 |
412 |
480 |
3542,844 696 |
|
Ni |
1608 |
3500-3600 |
1210-1320 |
1107 |
3334 |
410 |
481 |
3540, 853 696 |
|
Cu |
1608 |
3500-3600 |
1210-1320 |
1108 |
3342 |
420 |
485 |
3548, 833 696 |
Elemental Analysis
The elemental analysis data was found to corroborate the [C20H40N2O7M]Cl2 composition of the complexes and to be in conformity with suggested formulae. The complexes that are created have distinct hues from the ligand, which indicates that complexes are being formed. Complexation is supported by the melting temperatures of complexes, which are more than 250°C and distinct from those of free ligands. At room temperature, the produced complexes are stable, non-deliquescent, and non-hygroscopic. Various polar and non-polar solvents were used to test the solubility of complexes. All of the complexes are soluble in DMSO and DMF but insoluble in carbon tetrachloride, acetone, chloroform, ethyl alcohol, and water. All of the produced complexes appear to be monomers based on the analytical results and the complexes’ solubility characteristics. Table 2 showed the findings of the CHNO elemental analysis along with some physical characteristics of the ligand and its complexes.
Table 2: CHNO elemental analysis results and some physical properties of the Ligand and its complexes
|
Sample Formula |
M.Wt |
Colour |
M.P(oC) |
%Yield |
Elemental Analysis % Calc. |
|||
|
C |
H |
N |
O |
|||||
|
PGB |
159.23 |
White |
136 |
— |
60.29 |
10.67 |
8.79 |
20.09 |
|
TRB |
225.28 |
White |
247 |
— |
63.92 |
42.80 |
6.21 |
21.30 |
|
(A) |
549.25 |
|
>250 |
78% |
43.61 |
7.26 |
2.54 |
20.35 |
|
(B) |
550.13 |
|
>250 |
70% |
43.62 |
7.27 |
2,54 |
20.35 |
|
(C) |
554.98 |
|
>250 |
75% |
43.24 |
7.20 |
2.52 |
20.18 |
Mass Spectrometry
The mass spectra of the metal complexes were obtained using electron impact fragmentation. For the purpose of recovering large fragments associated with breakdown products and their complexes, high resolution mass spectrometry was typically employed29. The mass spectrum of the Co(II) complex (A) is displayed in Figure 3. This complex’s expected molecular weight is 549.25 g/mol for Co(II). The characteristic peaks at 170, 152, and 227 m/z could be caused by other fragments. The pieces’ stability is shown by their brilliance. Figure 3 displays the Ni (II) complex’s mass spectrum (B). A peak at 550.28 m/z was displayed by the complex moiety C20H40N2O7NiCl2 in the spectrum, matching the complex moiety’s peak. Different sections may be responsible for the 459, 362 and 312 m/z peaks at different frequencies. As illustrated in Figure 3, the mass spectrum of the Cu(II) complex (C). C20H40N2O7CuCl2 was identified as the chemical moiety by looking for a peak in the spectra at 554 m/z. The typical peaks at 170, 152, and 227 m/z could be caused by other factors.
 |
Figure 3: Mass spectrum of metal (A) Co(II), (B) Ni(II), and Cu(II) (C) complexes. |
Molecular Modeling
Our molecular docking analysis, conducted using the Autodock Vina PyRx tool, aimed to elucidate the interactions between metal complex compounds and four distinct protein targets associated with neurodegenerative diseases. The proteins under investigation include SLAS-micelle bound alpha-synuclein (PDB id: 2KKW), Alzheimer’s amyloid beta-peptide (PDB id: 1IYT), Alzheimer’s A beta (1-42) fibrils (PDB id: 2BEG), and micelle-bound human alpha-synuclein (PDB id: 1XQ8), which is implicated in the aetiology of Parkinson’s disease.
As illustrated in Table 3-6, our findings reveal compelling results regarding the binding energies and hydrogen bond interactions of our chemical compounds with the target proteins.
According to the data presented in Tables 3-6, our metal complex compounds exhibited notable binding energies and engaged in hydrogen bond interactions with key residues of the target proteins. Compound C, in particular, demonstrated consistently strong binding energies across multiple protein targets. The hydrogen bond interactions observed underscore the specificity of the interactions, contributing to the stability of the protein-ligand complexes.
These findings offer important new information about the manufactured metal complexes’ possible therapeutic value in relation to neurodegenerative illnesses. These intriguing computational results need to be confirmed and expanded upon by additional in vitro research and experimental validations.
Table 3: shown the interaction between PDB 1IYT and Metal complex
|
Sr, No. |
Compound Name |
Energy of Binding Kcal/mol |
Hydrogen-bond interaction |
Distance Ǻ |
|
1 |
A |
-5.4 |
GLU3 |
3.38 |
|
2 |
B |
-5.3 |
GLU3 |
2.42 |
|
3 |
C |
-5.2 |
HIS6 GLU3 |
2.66 2.26, 2.59 & 3.32 |
The molecular docking results demonstrated favorable binding energies for compounds A, B, and C with PDB 1IYT, indicating stable interactions. Notably, hydrogen bond interactions were observed, with Compound B displaying the closest interaction with GLU3 at 2.42 Å.
Table 4: shown the interaction between PDB 1XQ8 and Metal complex
|
Sr, No. |
Compound Name |
Energy of Binding Kcal/mol |
Hydrogen-bond interaction |
Distance Ǻ |
|
1 |
A |
-6.0 |
LYS45 LYS43 GLU35 |
2.38 2.37 2.64 |
|
2 |
B |
-5.9 |
VAL48 |
2.63 |
|
3 |
C |
-5.8 |
LYS |
2.49 |
In the case of PDB 1XQ8, compounds A, B, and C showcased robust binding energies, with Compound A exhibiting the lowest binding energy of -6.0 Kcal/mol. Hydrogen bond interactions were observed, emphasizing the specificity of interactions with key residues such as LYS45, LYS43, and GLU35.
Table 5: shown interaction between PDB 2BEG and Metal complex
|
Sr, No. |
Compound Name |
Energy of Binding Kcal/mol |
Hydrogen-bond interaction |
Distance Ǻ |
|
1 |
A |
-5.7 |
PHE19 |
2.58 |
|
2 |
B |
-5.9 |
ALA21 PHE19 |
2.23 & 2.56 2.59 |
|
3 |
C |
-6.1 |
ASP23 ASN27 LYS28 |
2.95 2.74 2.04 |
The interaction with PDB 2BEG revealed noteworthy binding energies for compounds A, B, and C. Hydrogen bond interactions were observed, with Compound C engaging multiple residues (ASP23, ASN27, LYS28), indicating its potential efficacy in binding to different regions of the protein.
Table 6: shown interaction between PDB 2KKW and Metal Complex
|
Sr, No. |
Compound Name |
Energy of Binding Kcal/mol |
Hydrogen-bond interaction |
Distance Ǻ |
|
1 |
A |
-5.0 |
LYS6 |
2.77, 2.76 & 2.20 |
|
2 |
B |
-5.3 |
LYS97 GLN99 GLY101 |
3.03 3.19 & 2.54 2.94 |
|
3 |
C |
-5.0 |
GLY101 |
2.15 |
The interaction with PDB 2KKW showcased competitive binding energies for compounds A, B, and C. Compound B displayed interactions with key residues LYS97, GLN99, and GLY101, emphasizing its potential in modulating protein function.
The observed favorable binding energies and specific hydrogen bond interactions across multiple proteins, as outlined in Tables 3-6, highlight the potential of our metal complexes to modulate the molecular events implicated in neurodegenerative pathways. Compound C, in particular, consistently demonstrated strong binding affinities and engaged in specific interactions with crucial residues, indicating its potential as a multifaceted therapeutic agent.
Overall, the consistent favorable binding energies and specific hydrogen bond interactions across multiple proteins suggest that our synthesized metal complex compounds hold promise as potential candidates for therapeutic interventions in neurodegenerative diseases. These computational findings lay the groundwork for further experimental validations, including in vitro studies, to confirm and extend the observed interactions and assess the practical implications of these metal complexes in a biological context.
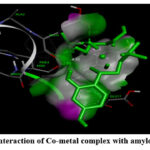 |
Figure 4: (A). Interaction of Co-metal complex with amyloid beta-peptide |
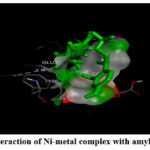 |
Figure 4: (B) Interaction of Ni-metal complex with amyloid beta-peptide. |
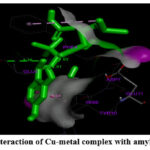 |
Figure 4: (C). Interaction of Cu-metal complex with amyloid beta-peptide |
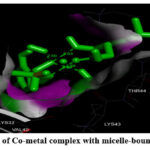 |
Figure 5: (A). Interaction of Co-metal complex with micelle-bound human alpha-synuclein |
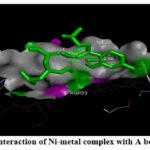 |
Figure 5: (B). Interaction of Ni-metal complex with micelle-bound human alpha-synuclein. |
 |
Figure 5: (C). Interaction of Cu-metal complex with micelle-bound human alpha-synuclein. |
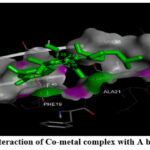 |
Figure 6: (A) Interaction of Co-metal complex with A beta (1-42) fibrils. |
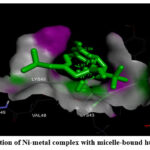 |
Figure 6: (B). Interaction of Ni-metal complex with A beta (1-42) fibrils. |
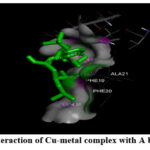 |
Figure 6: (C) Interaction of Cu-metal complex with A beta (1-42) fibrils. |
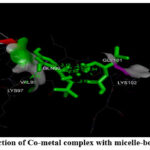 |
Figure 7: (A). Interaction of Co-metal complex with micelle-bound alpha-synuclein. |
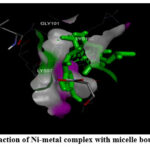 |
Figure 7: (B): Interaction of Ni-metal complex with micelle bound alpha-synuclein. |
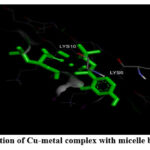 |
Figure 7: (C). Interaction of Cu-metal complex with micelle bound alpha-synuclein |
Conclusion
The study revealed that Terbutaline (TRB) functions as a bidentate ligand, offering two donating centers—one nitrogen atom from an amine group and one oxygen atom from a hydroxyl group. Furthermore, Pregabalin (PGB) displayed bidentate ligand behavior, coordinating with metal ions (Cu(II), Ni(II), and Co(II)) through a nitrogen atom from an amine group and an oxygen atom from a carboxylic group, resulting in the formation of novel mononuclear complexes. Employing diverse analytical and spectroscopic instruments, we characterized the metal complexes, confirming their coordination number as 6 and supporting the proposed octahedral structure. Molecular docking analysis elucidated that the binding energies of the metal complexes with proteins 1IYT, 2BEG, 1XQ8, and 2KKW ranged from -5 to -6 kcal/mol. Particularly noteworthy were the promising interactions observed, especially with 2BEG and 1XQ8, suggesting significant affinities. These findings underscore the potential therapeutic relevance of the developed metal complexes in the context of neurodegenerative disorders. While these computational insights provide a foundation, further research and experimental validations are imperative to advance these metal complexes as potential therapeutic candidates. The comprehensive investigation, encompassing interactions with proteins linked to Alzheimer’s and Parkinson’s diseases, sets the stage for subsequent in vitro studies. The integration of computational findings with empirical evidence will be pivotal in propelling the translational potential of these metal complexes toward effective treatments for neurodegenerative disorders.
Acknowledgement
The authors are grateful to the Sophisticated Analytical Instrument Facility, CIL Panjab University, Chandigarh and Sardar Patel University, Anand, Gujarat, India, for providing the facility and support for this work.
Conflict of interest
The author declares no conflict of interest to disclose.
Ethics of human and animal experiments
None
References
- Froestl, W., P. Yogeeswari, J. Ragavendran, D. Sriram, N. Bowery, D. Hill, A. Hudson et al. “Asymmetric Synthetic Strategies of (R)-(–)-Baclofen: An Antispastic Drug.” Synthesis 50, no. 02 (2018): 211-226.
CrossRef - Rose, M. A., and P. C. A. Kam. “Gabapentin: pharmacology and its use in pain management.” Anaesthesia 57, no. 5 (2002): 451-462.
CrossRef - Van Boven, J. F. M., E. G. Hiddink, A. G. G. Stuurman-Bieze, C. C. M. Schuiling-Veninga, M. J. Postma, and S. Vegter. “The pharmacists’ potential to provide targets for interventions to optimize pharmacotherapy in patients with asthma.” International journal of clinical pharmacy 35 (2013): 1075-1082.
CrossRef - Bastiansen, Anders, Sarah Eggert, and Erland Pedersen. “Pharmacokinetics of terbutaline in chronic kidney disease.” European journal of clinical pharmacology 69 (2013): 1951-1954.
CrossRef - Iftikhar, Imran H., Muhammad Imtiaz, Allan S. Brett, and David J. Amrol. “Cardiovascular safety of long acting beta agonist-inhaled corticosteroid combination products in adult patients with asthma: a systematic review.” Lung 192 (2014): 47-54.
CrossRef - Ali, Ahmed Mahmoud Abdelhaleem, and Mohamed Emam A. Abdelrahim. “Modeling and optimization of terbutaline emitted from a dry powder inhaler and influence on systemic bioavailability using data mining technology.” Journal of Pharmaceutical Innovation 9 (2014): 38-47.
CrossRef - Wade, Alexander, and Christopher Chang. “Evaluation and treatment of critical asthma syndrome in children.” Clinical reviews in allergy & immunology 48 (2015): 66-83.
CrossRef - Li, Shuting, Janshi Wang, and Shulin Zhao. “Determination of terbutaline sulfate by capillary electrophoresis with chemiluminescence detection.” Journal of Chromatography B 877, no. 3 (2009): 155-158.
CrossRef - Li, Yamin, Zhuo Ye, Jing Zhou, Jie Liu, Ge Song, Kai Zhang, and Baoxian Ye. “A new voltammetric sensor based on poly (L-arginine)/graphene–Nafion composite film modified electrode for sensitive determination of Terbutaline sulfate.” Journal of Electroanalytical Chemistry 687 (2012): 51-57.
CrossRef - Izquierdo-Lorenzo, Irene, Santiago Sánchez-Cortés, and José Vicente García-Ramos. “Adsorption of beta-adrenergic agonists used in sport doping on metal nanoparticles: a detection study based on surface-enhanced Raman scattering.” Langmuir 26, no. 18 (2010): 14663-14670.
CrossRef - Harris, Robin K., Paul Hodgkinson, Tomas Larsson, Amsaveni Muruganantham, Ingvar Ymén, Dmitry S. Yufit, and Vadim Zorin. “Characterization of polymorphs and solvates of terbutaline sulfate.” Crystal Growth and Design 8, no. 1 (2008): 80-90.
CrossRef - Selkoe, Dennis J., and John Hardy. “The amyloid hypothesis of Alzheimer’s disease at 25 years.” EMBO molecular medicine 8, no. 6 (2016): 595-608.
CrossRef - Gnanaraj, Charles, Mahendran Sekar, Shivkanya Fuloria, Shasank S. Swain, Siew Hua Gan, Kumarappan Chidambaram, Nur Najihah Izzati Mat Rani et al. “In silico molecular docking analysis of karanjin against alzheimer’s and parkinson’s diseases as a potential natural lead molecule for new drug design, development and therapy.” Molecules 27, no. 9 (2022): 2834.
CrossRef - Sun, Raymond Wai-Yin, Dik-Lung Ma, Ella Lai-Ming Wong, and Chi-Ming Che. “Some uses of transition metal complexes as anti-cancer and anti-HIV agents.” Dalton Transactions 43 (2007): 4884-4892.
CrossRef - García-Valverde, Maria, and Tomás Torroba. “Sulfur-nitrogen heterocycles.” Molecules 10.2 (2005): 318-320.
CrossRef - Gamil, Manar A., Sadeek A. Sadeek, Wael A. Zordok, and Walaa H. El-Shwiniy. “Spectroscopic, DFT modeling and biological study of some new mixed ligand metal complexes derived from gatifloxacin and pregabalin.” Journal of Molecular Structure 1209 (2020): 127941.
CrossRef - Crescenzi, Orlando, Simona Tomaselli, Remo Guerrini, Severo Salvadori, Anna M. D’Ursi, Piero Andrea Temussi, and Delia Picone. “Solution structure of the Alzheimer amyloid β‐peptide (1–42) in an apolar microenvironment: Similarity with a virus fusion domain.” European journal of biochemistry 269, no. 22 (2002): 5642-5648.
CrossRef - Ulmer, Tobias S., Ad Bax, Nelson B. Cole, and Robert L. Nussbaum. “Structure and dynamics of micelle-bound human α-synuclein.” Journal of Biological Chemistry 280, no. 10 (2005): 9595-9603.
CrossRef - Lührs, Thorsten, Christiane Ritter, Marc Adrian, Dominique Riek-Loher, Bernd Bohrmann, Heinz Döbeli, David Schubert, and Roland Riek. “3D structure of Alzheimer’s amyloid-β (1–42) fibrils.” Proceedings of the National Academy of Sciences 102, no. 48 (2005): 17342-17347.
CrossRef - Rao, Jampani Nageswara, Christine C. Jao, Balachandra G. Hegde, Ralf Langen, and Tobias S. Ulmer. “A combinatorial NMR and EPR approach for evaluating the structural ensemble of partially folded proteins.” Journal of the American Chemical Society 132, no. 25 (2010): 8657-8668.
CrossRef - https://chemdraw-ultra.software.informer.com/11.0/
CrossRef - https://discover.3ds.com/discovery-studio-visualizer-download
CrossRef - Trott, Oleg, and Arthur J. Olson. “AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading.” Journal of computational chemistry 31, no. 2 (2010): 455-461.
CrossRef - Dallakyan, Sargis, and Arthur J. Olson. “Small-molecule library screening by docking with PyRx.” Chemical biology: methods and protocols (2015): 243-250.
CrossRef - Yasin, Haya, Bashar Al-Taani, and Mutaz Sheikh Salem. “Preparation and characterization of ethylcellulose microspheres for sustained-release of pregabalin.” Research in Pharmaceutical Sciences 16, no. 1 (2021): 1.
CrossRef - Sreejan, M., V. Krishnaveni, Satyavathi K. Sai Padmini, P. Bhojaraju, and M. Madhubabu. “Design and evaluation of chronotherapeutics delivery of terbutaline sulphate by pulsincap technology.” Int J Drug Deliv Technol 7, no. 1 (2017): 75-82.
CrossRef - Al-Hamdani, Abbas Ali Salih, and Rehab Ghalib Hamoodah. “Transition metal complexes with tridentate ligand: preparation, spectroscopic characterization, thermal analysis and structural studies.” Baghdad Science Journal 13, no. 4 (2016): 0770-0770.
CrossRef - Al-Hamdani, Abbas Ali Salih, Najdat R. Al-Khafaji, and Naser Shaalan. “Preparation, spectral, thermal and bio activity studies of azo dyes complexes.” RESEARCH JOURNAL OF PHARMACEUTICAL BIOLOGICAL AND CHEMICAL SCIENCES 8, no. 3 (2017): 740-750.
- Suleman, Veyan Taher, Abbas Ali Salih Al‐Hamdani, Suzan Duraid Ahmed, Vian Yamin Jirjees, Mohammad Ehtisham Khan, Adnan Dib, Wail Al Zoubi, and Young Gun Ko. “Phosphorus Schiff base ligand and its complexes: Experimental and theoretical investigations.” Applied Organometallic Chemistry 34, no. 4 (2020): e5546.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









