Design, Synthesis, Molecular Docking, Invitro Anticancer and Antibacterial Evaluation of Novel Pyrazole Linked with Quinazoline Scaffolds
1Department of Pharmaceutical Chemistry, GITAM School of Pharmacy, GITAM (Deemed to be University), Sangareddy, Telangana, India.
2GITAM School of Pharmacy, GITAM (Deemed to be University), Sangareddy, Telangana, India.
Corresponding Author E-mail: mohdafroz1992@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/390635
Article Received on : 13 Jul 2023
Article Accepted on :
Article Published : 22 Nov 2023
Reviewed by: Dr. Tejeswara Rao Allaka
Second Review by: Dr. layla othman
Final Approval by: Dr. S. A Iqbal
A novel series of compounds are synthesized N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-oxo-3-(3,5-diphenyl-2H-pyrazol-1(5 H)-yl) propenamide (3a-l). All the synthesized compounds are characterized by different spectral tools 1HNMR, IR,13CNMR, and MASS. It was screened as in vitro anticancer and antibacterial activity. Among the synthesized compounds 3d and 3e exhibit potent against three cancer cell-line MCF7, PC-3, HT-29. IC50(µM) 3d (16.52, 13.24, 10.15 μg/ml) 3e (17.28, 15.26, 12.33 μg/ml) with standard drugs doxorubicin (15.29, 12.26, 9.06 μg/ml) and 5-fluorouracil (16.15, 13.73, 10.25 μg/ml). Antibacterial activity 3d, 3e, 3j, 3k scaffolds exhibit a promising activity with the standard drug ciprofloxacin. Insilico molecular docking is examined, Its predicted a good binding affinity against with 5C5S, 6XXN, 3K46 proteins.
KEYWORDS:Anti-cancer; Anti-bacterial; Molecular docking; Pyrazole; Quinazoline
Download this article as:| Copy the following to cite this article: Afroz M, kumar G. S. Design, Synthesis, Molecular Docking, Invitro Anticancer and Antibacterial Evaluation of Novel Pyrazole Linked with Quinazoline Scaffolds. Orient J Chem 2023;39(6). |
| Copy the following to cite this URL: Afroz M, kumar G. S. Design, Synthesis, Molecular Docking, Invitro Anticancer and Antibacterial Evaluation of Novel Pyrazole Linked with Quinazoline Scaffolds. Orient J Chem 2023;39(6). Available from: https://bit.ly/46szAwf |
Introduction
One of the most common aberrant cell growths in the human body is cancer. Death rates from cancer exist. If cancer is detected in its early stages, it can be cured; if it is detected in its later stages, there are few possibilities that it can be cured.1
Heterocyclic chemistry becomes more challenging for medicinal chemists. Heterocyclic compounds and their derivatives show various pharmacological and clinically active. Heterocyclic molecules have diversified action in various pharmacological and therapeutic activities.2 The organic molecules are distributed in nature and play an essential role in regulating biological processes. Fused heterocyclic moieties like pyrazole and quinazoline nucleus are significant in the drug discovery process The heterocyclic compounds like pyrazole,3 quinazoline4 nucleus contain prominent electron withdrawing groups like nitrogen and oxygen atom. They have the potency to bind the receptor and show an agonist effect to receptor and it exhibit molecular inhibitory binding with biological systems. Due to their privilege and efficacy towards pharmacological significance. It has wide-linked properties of Pyrazole and quinazoline nucleus has a key role of inhibiting certain receptors like aurora kinase inhibitors 5 cyclin-dependent kinases (CDK) inhibitors,5 reticular activating system-neuroendocrine tumor (Ras-Net),5 DNA binding agent,5 epithelial growth factor receptor (EPGR)6 inhibitor, cyclo-oxygenase(COX),7 lipoxygenase(LOX) 7 inhibitors and tubulin inhibition polymerization 8 further active inhibitory receptor is represented in Figure1.9 It attach to different hetero molecules scaffolds published in the literature data for the potency compounds has with a wide range of therapeutic properties. Pyrazole10 and quinazoline11 core moiety exhibit diverse pharmacological and therapeutic properties such as anticancer,12 anti-mycobacterial,13 antidiabetic,14 anti-inflammatory,15 antimicrobial,16 anti-leishmanial,17 anti-hypertensive agents α-blocking,18 Neuroprotective agents18 as approved by USFDA drugs are shown in Figure2
 |
Figure 1: Importance of inhibitory activity of Pyrazole and Quinazoline nucleus. |
 |
Figure 2: USFDA-approved drugs Pyrazole and Quinazoline nucleus. |
Extremely efforts to develop a synthesis of pyrazole linked with quinazoline and its scaffolds. In our research, grateful to these pyrazole and quinazoline hybrids in continuation of our work towards biologically active molecules.19 In this research our aim to synthesized newly novel pyrazole and quinazoline hybrids via Schiff mechanisms and mannish base mechanism and its scaffolds (3a-l). Synthesized scaffolds are promising against cancer cell lines MCF-7, PC-3, and HT-29 by using MTT assay methods. Antibacterial activity is screened by using the agar-diffusion method and insilico molecular docking 20 is performed.
Materials and Methods
All the chemicals were purchased from SD Fine and AURA Laboratories Analytical research grades. Then further characterization by different analytical techniques like silica gel (TLC) Thin layer chromatography and IR spectroscopy (BRUKER), 1H-NMR spectroscopy (400 MHz AvanceCore), 13C spectroscopy (Avance 400 MHz), Mass spectrometry (Thermo fisher scientific Orbitrap Exploris 120) and Melting point (LB-MPS8). Anti-cancer activity were performed by using MTT assay and antibacterial activity were performed by using bacterial strain and in silicomolecular docking are predicted.
Synthesis of N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-oxo-3-(3,5-diphenyl-2H-pyrazol-1(5 H)-yl) propanamide.
The procedure consist of three steps
Step I: Synthesis of Chalcones
Acetophenone and different substituted benzaldehydes an add ethanol Stirr at magnetic stirrer at room temperature1hr by using Claisen– Schmidt condensation mechanism and the reaction was observed in TLC. The reaction crude product was filter and recrystallized with ethanol and finally obtained a chalcone moiety.
Step II: Synthesis of Quinazoline malonic dihydrazide
Take Malonic di-hydrazide and add 2-methyl benzoxazine-4-one and add pyridine and refluxed for 3hr. In reaction mixture add crush ice or cold water neutralised with dil HCl. Filter the crude product and Precipitate were collected, dry the obtained product and it was recrystallized with methanol or ethanol.
Step III: Synthesis of Pyrazole Scaffolds (3a-3l)
Chalcone of step1 and malonic-dihydrozido quinazoline step2 was taken in RBF and add glacial acetic acid and add NaOH (40% 10 ml) then reflux for 5hr. The reaction undergo cyclization schiff and mannish base mechanism formation of pyrazole and its scaffold (3a-l). The progress of the reaction mixture was monitored in TLC cooled with rinse water and add crushed ice. Filter the reaction and obtain solid residue, dry the residue, and recrystallized with ethanol.
Biological activity evaluation
In-vitro Anti-cancer activity
Anti-cancer activity were evaluated (3a-l) against three cancer cell line breast cancer (MCF7) and prostate cancer (PC-3) and human colon cancer (HT-29) by using MTT assay method 21, Trypsinization of the cells and trypan blue assay were performed to determine the viability of the cells in suspension. Trypsinization of the cells and trypan blue assay were performed to determine the viability of the cells in suspension. The cells were then counted using a hemocytometer (IMPROVED B.S.748 NEUBAUER by Rohem india, and a density of 5.0 X 103 cells per well were seeded 100µL in culture medium in 96 well plates, and incubated Manufacturer Thermo scientific Model Co2 incubator model 370 manual for 12hrs at 370C. Removed the existing medium, then filled each well with new media. Following that, plates were incubated for three hours at 370C. Precipitations were produced at the end of the incubation time as a result of the MTT salt being reduced to chromophore-Formosan crystals by cells with metabolically active mitochondria. The optical density of crystals that had been dissolved in DMSO was calculated at 570 nm on a microplate reader using a FlexStation3 plate reader. The standard formula for calculating cell toxicity is (average OD*100/positive control)-100, and figures for the concentration of the test drug needed to hinder cell development by 50% (IC50) were utilized.
In-vitro Antibacterial activity
Antibacterial activity was evaluated (3a-l) against gram-positive and gram-negative bacterial strains by using agar plate diffusion method.22 It is examined that the view a dose-dependent inhibition of bacterial growth.
Insilico Molecular docking studies
The insilico molecular docking studies were predicted in Autodock vina PyRx 0.8.23 This tool was analyze and predict the ligand and completely bound to the receptor. Viewed in 3D structure. The protein was downloaded from WWW.RCSBPDB.COM (23) PDB: 5C5S (Human-myosin9b Rhogap), 6XXN (sting CTD complex), 3K46 (E.Coli beta glucurinadase) and finally docking visualization analysis was predicted in Discovery Studio and Molegro Molecular Viewer.
Spectral data of newly synthesized compound
3a: N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-oxo-3-(3,5-diphenyl-2H-pyrazol-1(5 H)-yl) propanamide.
IR: 3413(-NH), 3377(-NH), 3134(-CH), 2989(-CH3),2892(-CH),1725 (C=O), 1700 (C=O), 1555(C=N), 1295(C-N). 1H NMR: 12.1 (1H, -NH), 11.33(1H, -NH), 8.4- 6.9(13H Ar), 6.35 (1H, -CH), 3.2 (1H, -CH2), 2.42(3H, -CH3). 13C NMR 127.4, 133.5, 12.4, 147.1, 120.9, 128.8, 164, 161, 19.9, 170.3, 44.5, 95.3, 61, 134, 128.7, 128.0, 128.7, 126.4 MASS (ESI) m/z: 466 [M+H]+
3b: N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-oxo-3-(5-phenyl -3-p-tolyl-2H-pyrazol-1(5 H)-yl) propanamide.
IR: 3455 (-NH), 3302(-NH), 3102(-CH), 2957(-CH),1700 (C=O), 1670 (C=O), 1585(C=N), 1252(C-N).1H NMR: 12.5 (1H, -NH,), 11.3(1H, -NH), 8.0- 6.9(13H Ar), 6.83 (1H, -CH), 3.42 (1H, -CH2), 2.34 (3H, -CH3). 1.90 (3H, -CH3). 13C NMR 127.4, 133.5, 12.4, 147.1, 120.9, 128.8, 164, 161, 19.9, 170.3, 44.5, 95.3, 61, 134, 128.7, 128.0, 128.7, 126.4, 19.9, 24.3 MASS (ESI) m/z: 478 [M+H]+
3c: 3-(3-(4-methoxyphenyl)-5-phenyl-2H –pyrazol-1(5 H)-yl-N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-oxopropanamide.
IR: 3395 (-NH), 3355(-NH), 3144(-CH), 2980(-CH3),2895(-CH),1715 (C=O), 1710 (C=O), 1545(C=N), 1285(C-N), 1245(C-O-C-OCH3).1H NMR: 11.9 (1H, -NH), 10.3 (1H, -NH), 8.0- 6.9 (13H Ar), 6.55 (1H, -CH), 3.4 (1H, -CH2 ), 2.4 (3H, -CH3). 2.33 (3H, -OCH3). 13C NMR 127.4, 133.5, 12.4, 147.1, 120.9, 128.8, 164, 161, 19.9, 170.3, 44.5, 95.3, 61, 134, 128.7, 128.0, 128.7, 126.4, 159.9, 55.9 MASS (ESI) m/z: 496 [M+H]+.
3d: N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-(3-(4-nitrophenyl)-5-phenyl-2 H-pyrazol-1(5 H)-yl)-3-oxopropanamide.
IR: 3395 (-NH), 3365 (-NH), 3133(-CH), 2945(-CH3), 2900 (-CH),1710 (C=O), 1715 (C=O), 1590 (-NO2), 1550 (C=N), 1290 (C-N), 1H NMR: 12.0 (1H, -NH), 11.3 (1H, -NH), 8.0- 6.9 (13H Ar), 6.7 (1H, -CH), 3.0 (1H, -CH2), 2.10 (3H, -CH3) 13C NMR 127.4, 133.5, 12.4, 147.1, 120.9, 128.8, 164, 161, 19.9, 170.3, 44.5, 95.3, 61, 134, 128.7, 128.0, 128.7, 126.4, 127.3, 121.0 MASS (ESI) m/z: 511 [M+H]+.
3e: 3-(3-(4-chlorophenyl)-5-phenyl-2H –pyrazol-1(5 H)-yl-N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-oxopropanamide
IR: 3415 (-NH), 3385 (-NH), 3145 (-CH), 2960(-CH3),2890 (-CH),1710 (C=O), 1695 (C=O), 1570 (C=N), 1265 (C-N), 890 (C-Cl).1H NMR: 12.0 (1H, -NH), 11.2 (1H, -NH), 8.0- 6.8(13H), 6.25(1H, -CH), 3.2 (1H, -CH2), 2.2 (3H, -CH3) 13C NMR 127.4, 133.5, 12.4, 147.1, 120.9, 128.8, 164, 161, 19.9, 170.3, 44.5, 95.3, 61, 134, 128.7, 128.0, 128.7, 126.4, 133.5, 128.8, 127.8 MASS (ESI) m/z: 500 [M+H]+.
3f: 3-(3-(3,4-dimethoxyphenyl)-5-phenyl-2H –pyrazol-1(5 H)-yl-N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-oxopropanamide
IR: 3403 (-NH), 3333(-NH), 3160 (-CH), 2955 (-CH3), 2890 (-CH),1710 (C=O), 1705 (C=O), 1535 (C=N), 1295 (C-N), 1255 (C-O-C-OCH3).1H NMR: 12.2 (1H, -NH), 11.4 (1H, -NH), 8.00- 6.8 H), 6.2 (1H, -CH), 3.05 (1H, -CH2), 2.0(3H, -CH3). 2.0 (6H, -OCH3) 13C NMR 127.4, 133.5, 12.4, 147.1, 120.9, 128.8, 164, 161, 19.9, 170.3, 44.5, 95.3, 61, 134, 128.7, 128.0, 128.7, 126.4, 111.6, 128.8, 127.8 MASS (ESI) m/z: 526 [M+H]+.
3g: N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-oxo-3-(3-phenyl -5-p-tolyl-2H-pyrazol-1(5 H)-yl) propanamide.
IR: 3380 (-NH), 3345 (-NH), 3125 (-CH), 2975(-CH3), 2855 (-CH),1690 (C=O), 1700 (C=O), 1535 (C=N), 1240 (C-N).1H NMR: 11.2 (1H, -NH), 10.3 (1H, -NH), 8.0- 6.8 (13H Ar), 6.5 (1H, -CH), 3.6 (1H, -CH2), 2.45 (3H, -CH3), 1.7 (3H, -CH3) 13C NMR 127.4, 133.5, 12.4, 147.1, 120.9, 128.8, 164, 161, 19.9, 170.3, 44.5, 95.3, 61, 134, 128.7, 128.0, 128.7, 126.4, 24.3, 126.4, 128.7 MASS (ESI) m/z: 478 [M+H]+.
3h: N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-oxo-3-(3,5-di p-tolyl-2H-pyrazol-1(5 H)-yl) propanamide.
IR: 3385 (-NH), 3340 (-NH), 3105 (-CH), 2955 (-CH3), 2875 (-CH),1720 (C=O), 1715 (C=O), 1565 (C=N), 1235 (C-N).1H NMR: 12.3 (1H, -NH), 11.4 (1H, -NH), 8.0- 6.8 (13H, Ar), 6.3 (1H, -CH), 3.0 (1H, -CH2), 2.05(3H, -CH3), 1.9 (6H, -CH3) 13C NMR 127.4, 133.5, 12.4, 147.1, 120.9, 128.8, 164, 161, 19.9, 170.3, 44.5, 95.3, 61, 134, 128.7, 128.0, 128.7, 126.4, 126.3, 24.3, 24.3 Mass (ESI) m/z: 494 [M+H]+.
3i: 3-(3-(3,4-methoxyphenyl)-5-p-tolyl-2H –pyrazol-1(5 H)-yl-N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-oxopropanamide
IR: 3425 (-NH), 3390 (-NH), 3145 (-CH), 2930 (-CH3), 2880 (-CH),1720 (C=O), 1700 (C=O), 1570 (C=N), 1295 (C-N), 1235 (C-O-C-OCH3).1H NMR: 12.0 (1H, -NH), 11.1 (1H, -NH), 8.0- 6.7 (13H, Ar), 6.0 (1H, -CH), 3.0 (1H, -CH2), 2.40 (3H, -CH3), 2.0 (3H, -OCH3), 1.85 (3H, -CH3) 13C NMR 127.4, 133.5, 12.4, 147.1, 120.9, 128.8, 164, 161, 19.9, 170.3, 44.5, 95.3, 61, 134, 128.7, 128.0, 128.7, 126.4, 127.4, 24.3, 55.9 Mass (ESI) m/z: 510 [M+H]+.
3j: N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-(3-(4-nitrophenyl)-5- p-tolyl-2H-pyrazol-1(5 H)-yl) -3-oxopropanamide
IR: 3370 (-NH), 3337(-NH), 3140 (-CH), 2920 (-CH3), 2885 (-CH),1710 (C=O Str), 1700 (C=O), 1580(-NO2), 1555 (C=N), 1280 (C-N).1H NMR: 12.0 (1H, -NH), 11.2 (1H, -NH), 8.1- 6.8(13H, Ar), 6.5 (1H, -CH), 3.5 (1H, -CH2), 2.5 (3H, -CH3), 1.5 (3H, -CH3) 13C NMR 127.4, 133.5, 12.4, 147.1, 120.9, 128.8, 164, 161, 19.9, 170.3, 44.5, 95.3, 61, 134, 128.7, 128.0, 128.7, 126.4, 121, 127.3, 24.3 MASS (ESI) m/z: 530 [M+H]+.
3k: : 3-(3-(4-chlorophenyl)-5-p-tolyl-2H –pyrazol-1(5 H)-yl-N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-oxopropanamide
IR: 3375 (-NH), 3330 (-NH), 3110 (-CH), 2935 (-CH3), 2836 (-CH), 1700 (C=O), 1690 (C=O), 1555 (C=N), 1290 (C-N), 878 (C-Cl).1H NMR 6.3 (1H, -CH), 3.1 (1H, -CH2), 2.5 (3H, -CH3), 1.9 (3H, -CH3) 13C NMR 127.4, 133.5, 12.4, 147.1, 120.9, 128.8, 164, 161, 19.9, 170.3, 44.5, 95.3, 61, 134, 128.7, 128.0, 128.7, 126.4, 128.8, 127.8, 24.3 MASS (ESI) m/z: 514 [M+H]+.
3l: 3-(3-(3,4-dimethoxyphenyl)-5-p-tolyl-2H –pyrazol-1(5 H)-yl-N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-oxopropanamide
IR: 3385(NH) 3360 (NH) 3115 (CH) 2925 (CH3) 2889 (CH) 1700 (C=O), 1690 (C=O), 1555 (C=N), 1290 (C-N), 1225 (OCH3).1H-NMR: 12.2 (1H, -NH), 11.3 (1H, -NH), 8.2- 6.8 9(13H, Ar), 6.15(1H, -CH) 3.3 (1H, -CH2),2.5 (6H, -OCH3), 2.7 (3H, -CH3), 1.8 (3H, -CH3) 13C NMR 127.4, 133.5, 12.4, 147.1, 120.9, 128.8, 164, 161, 19.9, 170.3, 44.5, 95.3, 61, 134, 128.7, 128.0, 128.7, 126.4, 24.3, 56.2, 56.2 MASS (ESI) m/z: 540 [M+H]+.
Results and Discussion
Chemistry
The research is focused on the precise synthesis of novel N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-oxo-3-(3,5-diphenyl-2H-pyrazol-1(5 H)-yl) propanamide (3a-l). The step1 is synthesized chalcones by using Claisen–Schmidt condensation and step2 are synthesized Quinazoline malonic dihydrazide step3 undergoes cyclization Schiff and Mannish base mechanism formation of pyrazole and its scaffold (3a-l) and are confirmed by using different spectral analysis tools 1HNMR, 13CNMR, IR and Mass. The completed scheme is represented in Figure 3 and the Physical properties are shown in Table 1
 |
Figure 3: Scheme I of novel pyrazole linked with quinazoline scaffolds (3a-3l). |
Table 1: Physical properties of novel pyrazole linked with quinazoline scaffolds (3a-3l).
|
Compounds |
Molecular Formula |
Molecular Weight |
R |
R1 |
R2 |
Melting Point |
Yield % |
|
3a |
C27H23N5O3 |
465.15 |
H |
H |
H |
2460C |
78% |
|
3b |
C28H25N5O3 |
479.12 |
CH3 |
H |
H |
2870C |
88% |
|
3c |
C28H25N5O4 |
495.08 |
OCH3 |
H |
H |
3240C |
86% |
|
3d |
C27H22N6O5 |
510.07 |
NO2 |
H |
H |
2760C |
78% |
|
3e |
C27H22N6O3Cl |
499.9 |
Cl |
H |
H |
2450C |
84% |
|
3f |
C29H27N5O5 |
525.16 |
OCH3 |
OCH3 |
H |
3450C |
72% |
|
3g |
C28H25N5O3 |
479.11 |
H |
H |
CH3 |
2260C |
85% |
|
3h |
C29H27N5O3 |
493.02 |
CH3 |
H |
CH3 |
2450C |
78% |
|
3i |
C29H27N5O4 |
509.02 |
OCH3 |
H |
CH3 |
2800C |
90% |
|
3j |
C28H24N6O5 |
529.01 |
NO2 |
H |
CH3 |
2520C |
89% |
|
3k |
C28H24N5O3Cl |
513.18 |
Cl |
H |
CH3 |
2450C |
78% |
|
3l |
C30H29N5O5 |
539.13 |
OCH3 |
OCH3 |
CH3 |
3150C |
82% |
Biological Activity
In-vitro Anticancer Activity
In our research. A Novel series of compounds are synthesized and evaluated as in-vitro anticancer activity (3a-l) were assessed against three cell lines (MCF7) and (PC-3) and (HT-29) by using MTT assay method 3d and 3e were found potent against all the three cell lines IC50(µM) 3d (16.52,13.24.10.15 μg/ml) 3e (17.28, 15.26, 12.33 μg/ml) compared with standard drug Doxorubicin (15.29, 12.26, 9.06 μg/ml) and 5-Fluorouracil (16.15, 13.73, 10.25 μg/ml). The IC50 values are represented in Table 2 and the standard graph is represented in Figure 4
Table 2: Anticancer activity of novel pyrazole linked with quinazoline scaffolds IC50 values (3a-3l).
|
COMPOUNDS |
IC50 (µM) |
||
|
MCF-7 |
PC-3 |
HT-29 |
|
|
3a |
39.26 ± 0.32 |
34.32 ± 0.41 |
31.26 ± 0.49 |
|
3b |
45.59 ± 0.51 |
62.18 ± 0.25 |
58.15 ± 0.16 |
|
3c |
95.23 ± 0.86 |
94.16 ± 0.48 |
92.23 ± 0.27 |
|
3d |
16.52 ± 0.052 |
13.24 ± 0.044 |
10.15 ± 0.032 |
|
3e |
17.28 ± 0.065 |
15.26 ± 0.072 |
12.33 ± 0.068 |
|
3f |
85.19 ± 0.16 |
90.69 ± 0.58 |
82.22 ± 0.32 |
|
3g |
46.02 ± 0.27 |
45.34 ± 0.32 |
43.26 ± 0.62 |
|
3h |
88.21 ± 0.48 |
85.26 ± 0.51 |
83.45 ± 0.38 |
|
3i |
35.12 ± 0.21 |
36.44 ± 0.64 |
33.19 ± 0.55 |
|
3j |
48.14 ± 0.71 |
42.14 ± 0.16 |
44.28 ± 0.67 |
|
3k |
47.25 ± 0.25 |
42.19 ± 0.56 |
39.15 ± 0.42 |
|
3l |
92.45 ± 0.68 |
87.21 ± 0.69 |
83.24 ± 0.22 |
|
DOXORUBICIN |
15.29 ± 0.032 |
12.26 ± 0.041 |
9.06 ± 0.028 |
|
5-FLUROURACIL |
16.15 ± 0.016 |
13.73 ± 0.025 |
10.25 ± 0.018 |
 |
Figure 4: Graphical representation of Anticancer activity of novel pyrazole linked with quinazoline scaffolds |
In-vitro Antibacterial activity
In our research in vitro Antibacterial activity was screened. The novel series of compounds are synthesized (3a-l) was examined against two gram-positive and two gram-negative bacteria strains by using the agar plate method. Interestingly, (3d, 3e,3j,3k) were potent and active against both the bacterial strains. The percentage zone of inhibition values are represented in Table 3 The results show that the substituted of 3d Nitrogen, 3e Chlorine, 3j Methyl and Nitrogen, 3k Methyl and Chlorine substituted compound shows a promising activity comparison with ciprofloxacin.
Table 3: Antibacterial activty of novel pyrazole linked with quinazoline scaffolds (3a-3l).
|
|
Gram-positive bacteria |
Gram-negative bacteria |
||||||
|
Staphylococcus Aureus |
Bacillus Subtilis |
Escherichia Coli |
Pseudomonas Aeruginosa |
|||||
|
Compounds |
50µg/ml |
100µg/ml |
50µg/ml |
100µg/ml |
50g/ml |
100µg/ml |
50µg/ml |
100µg/ml |
|
3a |
20 |
23 |
8 |
19 |
18 |
21 |
20 |
14 |
|
3b |
14 |
17 |
11 |
13 |
10 |
12 |
12 |
16 |
|
3c |
11 |
17 |
9 |
12 |
13 |
14 |
15 |
18 |
|
3d |
25 |
30 |
24 |
27 |
24 |
28 |
27 |
30 |
|
3e |
25 |
31 |
23 |
27 |
25 |
29 |
27 |
31 |
|
3f |
7 |
10 |
10 |
13 |
15 |
18 |
17 |
20 |
|
3g |
12 |
15 |
15 |
17 |
10 |
12 |
14 |
19 |
|
3h |
11 |
14 |
10 |
12 |
15 |
11 |
8 |
18 |
|
3i |
9 |
10 |
12 |
8 |
13 |
8 |
12 |
10 |
|
3j |
22 |
27 |
18 |
20 |
21 |
22 |
19 |
25 |
|
3k |
23 |
29 |
19 |
21 |
23 |
26 |
22 |
27 |
|
3l |
14 |
15 |
10 |
13 |
9 |
12 |
11 |
14 |
|
Ciprofloxacin |
28 |
33 |
25 |
29 |
27 |
32 |
30 |
34 |
Insilico Molecular docking
The insilico molecular docking was analyzed by using Autodock vina PyRx (0.8).The Autodock vina tool shows the accurate result, whenever software is run for trail the docking and binding affinity values are standard and accurate. The protein was downloaded from WWW.RCSBPDB.COM The protein and ligands are uploaded in PyRx tool and adjust coordinates of the x and y-axis. Then, This tool detects interaction with different amino acids and binding affinity values have been predicted in Excel file. Finally obtained binding affinity values and 2D and 3D images. It can be viewed by using tools like Discovery Studio and Molegro Molecular Viewer. The binding affinity values are represented in Table 4 and interaction with different amino acids is represented in figure 5 and 6
Table 4: Insilco molecular docking binding affinity values of novel pyrazole linked with quinazoline scaffolds (3a-3l).
|
BINDING -AFFINITY Kcal/mol |
|||
|
COMPOUNDS |
5C5S |
6XXN |
3K46 |
|
3a |
-10.3 |
-10.3 |
-11.2 |
|
3b |
-10.9 |
-10.7 |
-10.2 |
|
3c |
-10.5 |
-10.2 |
-10.5 |
|
3d |
-13.5 |
-12.8 |
-13.3 |
|
3e |
-12.8 |
-11.9 |
-12.6 |
|
3f |
-9.5 |
-10.8 |
-10.6 |
|
3g |
-10.9 |
-10.6 |
-11.4 |
|
3h |
-11.3 |
-10.8 |
-11.7 |
|
3i |
-10.9 |
-10.4 |
-10.7 |
|
3j |
-11.2 |
-10.8 |
-12.2 |
|
3k |
-11.1 |
-11.0 |
-11.8 |
|
3l |
-11.0 |
-10.1 |
-10 |
In our research. Insilico Molecular docking was performed and obtained good binding affinity values against PDB ID 5C5S (human myosin 9b RhoGAP), 6XXN (sting CTD complex), 3K46 (E.Coli beta glucurinadase). The binding affinity of the ligand 3d and 3e against proteins is around -12 kcal/mol. The ligand coupled with various amino acids Thr 112(A), Gln 108(A), Asn 139(A), Tyr 111(A), Gly 110(A), Ala 391(A) and Ile 143(A). The molecules binds to protein through its 2D and 3D complex. The protein and ligand bind and coupled with different amino acid interactions are completely moulded each other. In figure the ligand and protein mould through its 3D complex.
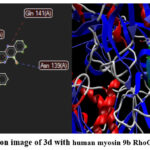 |
Figure 5: Interaction image of 3d with human myosin 9b RhoGAP (Pdb id – 5C5S) |
 |
Figure 6: Interaction image of 3e with E.Coli beta glucurinadase (Pdb id – 3k46). |
Conclusion
In this research, The Novel series of compounds are synthesized N-(2-methyl-4-oxoquinazolin-3(4H)-yl)-3-oxo-3-(3,5-diphenyl-2H-pyrazol-1(5H)-yl) propanamide (3a-l). The synthesized derivatives are characterized by 1HNMR, 13CNMR, IR, and MASS. It was evaluated as invitro anticancer and antibacterial activity. Among the series of compounds 3d and 3e exhibit potent against three cancer cell lines MCF7, PC-3, HT-29. IC50(µM) 3d (16.52, 13.24, 10.15 μg/ml) 3e (17.28, 15.26, 12.33 μg/ml) with standard drugs doxorubicin (15.29, 12.26, 9.06 μg/ml) and 5-fluorouracil (16.15, 13.73, 10.25 μg/ml). And also screened antibacterial activity by using agar plate diffusion methods 3d, 3e, 3j, 3k scaffolds exhibit a promising activity compared with the standard drug ciprofloxacin. In our research interestingly withdrawing group attached with scaffolds it shows a more active anti-cancer activity as well as antibacterial activity and also shows based on withdrawing capability. Insilico Molecular Docking performed and it is predicted a good binding affinity against 5C5S(Human-myosin9b-Rhogap), 6XXN (sting CTD complex), 3K46 (E.Coli beta glucurinadase) proteins.
Acknowledgement
The authors are grateful to the director of GITAM for Providing a Lab facility and IICT for providing spectral analysis.
Conflicts of Interest
There are no conflicts of interest declared by the authors.
References
- Nasir A, Bullo MM, Ahmed Z, Imtiaz A, Yaqoob E, Safdar M, Ahmed H, Afreen A, Yaqoob S. Nutrigenomics: Epigenetics and cancer prevention A comprehensive review. Crit Rev Food Sci Nutr. 2022 Mar 31;60(8):1375-87.
CrossRef - Gul S, Aslam K, Pirzada Q, Rauf A, Khalil AA, Semwal P, Bawazeer S, Al-Awthan YS, Bahattab OS, Al Duais MA, Thiruvengadam M. Xanthones: A class of heterocyclic compounds with anticancer potential Curr. Top. Med. Chem. 2022 Sep 1;22(23):1930-49.
CrossRef - Maciejewska N, Olszewski M, Jurasz J, Serocki M, Dzierzynska M, Cekala K, Wieczerzak E, Baginski M. Novel chalcone-derived pyrazoles as potential therapeutic agents for the treatment of non-small cell lung cancer. Sci. Rep. 2022 8;12(1):1-9.
CrossRef - Auti PS, Nandi A, Kumari V, Paul AT. Design, synthesis, biological evaluation and molecular modelling studies of oxoacetamide warhead containing indole-quinazolinone based novel hybrid analogues as potential pancreatic lipase inhibitors. New J. Chem. 2022;46(24):11648-61.
CrossRef - Kumar H, Saini D, Jain S, Jain N. Pyrazole scaffold: a remarkable tool in the development of anticancer agents. Eur. J. Med. Chem. 2013 1;70:248-58.
CrossRef - Al-Anazi M, Khairuddean M, Al-Najjar BO, Alidmat MM, Kamal NN, Muhamad M. Synthesis, anticancer activity and docking studies of pyrazoline and pyrimidine derivatives as potential epidermal growth factor receptor (EGFR) inhibitors. Arab. J. Chem. 2022 1;15(7):103864.
CrossRef - Gökhan-Kelekçi N, Koyunoğlu S, Yabanoğlu S, Yelekçi K, Özgen Ö, Uçar G, Erol K, Kendi E, Yeşilada A. New pyrazoline bearing 4 (3H)-quinazolinone inhibitors of monoamine oxidase: Synthesis, biological evaluation, and structural determinants of MAO-A and MAO-B selectivity. Bioorganic Med. Chem.. 2009 15;17(2):675-89.
CrossRef - Dwivedi AR, Rawat SS, Kumar V, Kumar N, Anand P, Yadav RP, Baranwal S, Prasad A, Kumar V. Synthesis and screening of novel 4-N-heterocyclic-2-aryl-6, 7, 8-trimethoxyquinazolines as antiproliferative and tubulin polymerization inhibitors. Bioorganic Med. Chem.2022 15;72:116976.
CrossRef - Afroz M, Kumar GS. Design Synthesis, Molecular Docking, and in vitro Anticancer and Antibacterial Evaluation of Novel 2-(9-chloro-2, 3-dimethyl-6, 7-dihydro-5H-benzo [7] annulen-8-yl)-3-thiazolidin-4one. J Young Pharm. 2023 Apr 17;15(2):283-91.
CrossRef - Al-Saheb R, Makharza S, Al-Battah F, Abu-El-Halawa R, Kaimari T, Abu Abed OS. Synthesis of new pyrazolone and pyrazole-based adamantyl chalcones and antimicrobial activity. Biosci. Rep. 2020 Sep 30;40(9).
CrossRef - Anh DT, Hai PT, Huy LD, Ngoc HB, Ngoc TT, Dung DT, Park EJ, Song IK, Kang JS, Kwon JH, Tung TT. Novel 4-oxoquinazoline-based N-hydroxypropenamides as histone deacetylase inhibitors: Design, synthesis, and biological evaluation. ACS omega. 2021 Feb 8;6(7):4907-20.
CrossRef - Thiriveedhi A, Nadh RV, Srinivasu N, Kaushal K. Novel hybrid molecules of quinazoline chalcone derivatives: Synthesis and study of in vitro cytotoxic activities. Lett Drug Des Discov. 2018 Jul 1;15(7):757-65.
CrossRef - Kumar A, Kumari N, Bhattacherjee S, Venugopal U, Parwez S, Siddiqi MI, Krishnan MY, Panda G. Design, synthesis and biological evaluation of (Quinazoline 4-yloxy) acetamide and (4-oxoquinazoline-3 (4H)-yl) acetamide derivatives as inhibitors of Mycobacterium tuberculosis bd oxidase. Eur. J. Med. Chem. 2022 15;242:114639
CrossRef - Azimi F, Azizian H, Najafi M, Hassanzadeh F, Sadeghi-Aliabadi H, Ghasemi JB, Faramarzi MA, Mojtabavi S, Larijani B, Saghaei L, Mahdavi M. Design and synthesis of novel quinazolinone-pyrazole derivatives as potential α-glucosidase inhibitors: Structure-activity relationship, molecular modeling and kinetic study. Bioorg. Chem. 2021 1;114:105127.
CrossRef - Mantzanidou M, Pontiki E, Hadjipavlou-Litina D. Pyrazoles and pyrazolines as anti-inflammatory agents. Molecules. 2021 Jan;26(11):3439.
CrossRef - Othman IM, Nasr HM, Hassan MI. Synthesis of some novel pyridazine, thienopyridazine, pyrazolopyridine, pyridopyrazolopyrimidine and pyridopyrazolotriazine derivatives with their antimicrobial activity. Can Chem Trans.2014;2(4):504-17..
CrossRef - Mowbray CE, Braillard S, Speed W, Glossop PA, Whitlock GA, Gibson KR, Mills JE, Brown AD, Gardner JM, Cao Y, Hua W. Novel amino-pyrazole ureas with potent in vitro and in vivo antileishmanial activity J. Med. Chem. 2015 Dec 24;58(24):9615-24.
CrossRef - Mathew B, Parambi DG, Mathew GE, Uddin MS, Inasu ST, Kim H, Marathakam A, Unnikrishnan MK, Carradori S. Emerging therapeutic potentials of dual‐acting MAO and AChE inhibitors in Alzheimer’s and Parkinson’s diseases. Archiv der Pharmazie. 2019 Nov;352(11):1900177.
CrossRef - Afroz M, Shiva Kumar G. Microwave-Assisted Synthesis, Molecular Docking Studies and Biological Evaluation of Novel Thiazole, Imidazole-Indole Hybrids. Asian J. Chem,2023; 35(3):705-11.
CrossRef - Sylvester PW. Optimization of the tetrazolium dye (MTT) colorimetric assay for cellular growth and viability. Lett Drug Des Discov. 2011 (pp. 157-168). Humana Press.
CrossRef - Zarrindokht EK, Pegah C. Antibacterial activity of ZnO nanoparticle on gram-positive and gram-negative bacteria Afr. j. microbiol. res. 2011 Jun 18;5(12):1368-73.
CrossRef - Kong R, Yi F, Wen P, Liu J, Chen X, Ren J, Li X, Shang Y, Nie Y, Wu K, Fan D. Myo9b is a key player in SLIT/ROBO-mediated lung tumor suppression. J. Clin. Investig. 2015, 125(12):4407-20.
CrossRef - Sahoo M, Jena L, Daf S, Kumar S. Virtual screening for potential inhibitors of NS3 protein of Zika virus. Genom. Inform.2016, 14(3):104.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.










