Design, In Silico Studies, and Synthesis of Some Azole Derivatives as Antimicrobial Agents
Syeda Huma Haider Zaidi1* , Abida Ash Mohd2
, Abida Ash Mohd2 , Mohd Imran2
, Mohd Imran2 , Menwah Khalifah Alshammari3, Khattab Fahed Alfrah3
, Menwah Khalifah Alshammari3, Khattab Fahed Alfrah3
1Department of Chemistry, College of Science, Northern Border University, Arar, Saudi Arabia.
2Department of Pharmaceutical Chemistry, College of Pharmacy, Northern Border University, Rafha, Saudi Arabia.
3College of Pharmacy, Northern Border University, Rafha, Saudi Arabia.
Corresponding Author E-mail: syeda.Zaidi@nbu.edu.sa
DOI : http://dx.doi.org/10.13005/ojc/390618
Article Received on : 06 Sep 2023
Article Accepted on :
Article Published : 06 Nov 2023
Reviewed by: Dr. Mani Vannan
Second Review by: Dr. N Raman
Final Approval by: Dr. Gavat Cristian
This work relates to the discovery of safer and more potent triazole-pyridazinone hybrid (TP) compounds as an inhibitor of sterol 14α-demethylase (SDM). The chemical structures of thirty-three TPs (TP1 to TP33) were designed. The docking scores (DS) of TPs were determined by molecular docking software utilizing three different proteins of SDM (PDB IDs: 3LD6, 5FSA, and 5TZ1). The ProTox II web server predicted TPs' oral LD50 and toxicity class (TC), whereas the Swiss-ADME database anticipated their pharmacokinetic parameters. Based on the in silico study data, four TPs (TP18, TP22, TP27, and TP33) were synthesized and evaluated for their in vitro antifungal activity against seven fungi. The DS (kcal/mol) of TP18 (3LD6 = -8.27; 5FSA = -9.07; 5TZ1 = -9.42), TP22 (3LD6 = -8.23; 5FSA = -8.93; 5TZ1 = -9.57), TP27 (3LD6 = -8.31; 5FSA = -9.12; 5TZ1 = -9.38), and TP33 (3LD6 = -8.19; 5FSA = -8.98; 5TZ1 = -9.94) were better than the DS of fluconazole (3LD6 = -8.18; 5FSA = -8.79; 5TZ1 = -9.18) and ketoconazole (3LD6 = -8.16; 5FSA = -8.86; 5TZ1 = -9.06) implying high potency of TP18, TP22, TP27 and TP33 than fluconazole and ketoconazole against SDM. The anticipated LD50 and toxicity class (TC) of TP18 (500 mg/kg; TC 4), TP22 (500 mg/kg; TC 4), TP27 (1000 mg/kg; TC 4), and TP33 (1000 mg/kg; TC 4) was better than ketoconazole (166 mg/kg; TC 3). The Swiss-ADME database results revealed that TP18, TP22, TP27, and TP33 passed Lipinski’s drug-likeliness rule and demonstrated high oral absorption and bioavailability comparable to ketoconazole and fluconazole. The synthesized compounds' spectral data (FTIR, 1H-NMR, 13C-NMR, and Mass) aligned to their designed chemical structure. The antifungal activity data implies that TP18, TP22, TP27, and TP33 were better antifungal agents than fluconazole and ketoconazole against tested fungi. These findings concurred with the DS of TP18, TP22, TP27, and TP33. In conclusion, TP18, TP22, TP27, and TP33 represent a new chemical template for developing safer and better SDM inhibitors as antifungal agents.
KEYWORDS:Azole; Antifungal activity Triazole; Molecular Docking; Pyridazinone; Synthesis
Download this article as:| Copy the following to cite this article: Zaidi S. H. H, Mohd A. A, Imran M, Alshammari M. K, Alfrah K. F. Design, In Silico Studies, and Synthesis of Some Azole Derivatives as Antimicrobial Agents. Orient J Chem 2023;39(6). |
| Copy the following to cite this URL: Zaidi S. H. H, Mohd A. A, Imran M, Alshammari M. K, Alfrah K. F. Design, In Silico Studies, and Synthesis of Some Azole Derivatives as Antimicrobial Agents. Orient J Chem 2023;39(6). Available from: https://bit.ly/40tdF6o |
Introduction
Antimicrobial resistance (AMR) is notoriously challenging and affects a patient’s quality of life in all aspects 1-3. The progress of AMR against the current antifungal drugs is also increasing 2. Discovering a new chemical template that can target essential enzymes of fungi (for example, sterol 14α-demethylase) is one of the promising research areas for developing safer and better antifungal agents 4,5. Sterol 14α-demethylase (SDM) is an established drug target to develop a new generation of antifungal drugs 6. SDM converts sterols to ergosterol in fungi, which is required for the proper cell membrane functions. Inhibition of SDM increases the permeability of the cell membrane due to a decline in the generation of ergosterol 6,7.
Azole is a general term for a 5-membered heterocyclic ring (imidazole, triazole, pyrazole, tetrazole, pentazole, oxazole, thiazole, and isoxazole) comprising at least one nitrogen and other hetero atoms 6-8. Many azole heterocycle-based SDM inhibitors, including triazole-based drugs (fluconazole, voriconazole, posaconazole, itraconazole, and isavuconazole) and imidazole-based drugs (ketoconazole, miconazole, and clotrimazole) are in clinical practice as antifungal agents 7,8. However, the development of the AMR is rising against azole heterocycle-based SDM inhibitors 2. The increasing incidences of azole-resistant SDM and the side effects (hepatotoxicity, allergic reactions, skin rashes, hormone-related effects, etc.) associated with the clinically used azole-based drugs are alarming to identify new, safer, and more effective azole-based antifungals 2. Recent studies have addressed pyridazinone-based heterocycles as appreciable antifungal agents 9-15. Pyridazinone and triazole derivatives have demonstrated antifungal activity 2,6-8,9-15. Accordingly, it was decided to identify non-toxic and potent triazole-pyridazinone hybrid molecules (TP) (Figure 1) as SDM inhibitors possessing promising pharmacokinetic parameters.
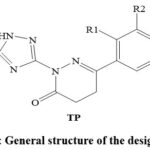 |
Figure 1: General structure of the designed TPs |
Experimental
General
The names of the software utilized in this research work have been mentioned in the relevant experimental parts. The chemicals and reagents used in this study were purchased from Sigma (USA). Spectral information on the synthesized compounds was collected using a variety of instruments, including a Gallenkamp melting point apparatus, a Shimadzu 440 spectrophotometer (for producing FTIR data), a Varian Gemini 500/125 MHz spectrophotometer (for producing 1H-NMR and 13C-NMR data, respectively), and a 70 eV GCMS/QP 1000 Ex mass spectrophotometer (for producing mass spectra).
Designing of TPs
The chemical structures of thirty-three compounds (TP1 to TP33) were designed based on the reaction between 3-hydrazineyl-5-(methylthio)-1H-1,2,4-triazole disclosed in the United States Patent Number US3331840A 16 and 4-oxobutanoic acid derivatives disclosed in the United States Patent Number US4052395A 17 (Figure 1) (Table 1).
Table 1: Chemical structures of the designed TPs (Figure 1).
|
Compound |
R1 |
R2 |
R3 |
R4 |
Compound |
R1 |
R2 |
R3 |
R4 |
|
TP1 |
H |
Cl |
CH3 |
Cl |
TP18 |
OH |
H |
CH3 |
H |
|
TP2 |
H |
Cl |
CH3 |
H |
TP19 |
OH |
H |
CH3 |
Cl |
|
TP3 |
H |
Br |
CH3 |
H |
TP20 |
OH |
Cl |
H |
Cl |
|
TP4 |
H |
Cl |
isopropyl |
H |
TP21 |
OH |
H |
Cl |
H |
|
TP5 |
H |
Br |
CH3 |
Br |
TP22 |
OH |
H |
H |
Cl |
|
TP6 |
H |
Cl |
ethyl |
Cl |
TP23 |
H |
H |
OCH3 |
Cl |
|
TP7 |
H |
Cl |
n-propyl |
Cl |
TP24 |
H |
Cl |
Cl |
H |
|
TP8 |
OH |
Cl |
CH3 |
Cl |
TP25 |
H |
Br |
CH3 |
Cl |
|
TP9 |
H |
H |
CH3 |
Cl |
TP26 |
H |
Cl |
OH |
Cl |
|
TP10 |
H |
H |
H |
Br |
TP27 |
H |
H |
NH2 |
H |
|
TP11 |
H |
H |
H |
Cl |
TP28 |
H |
Br |
NH2 |
H |
|
TP12 |
H |
Cl |
n-butyl |
Cl |
TP29 |
H |
H |
NHCOCH3 |
Cl |
|
TP13 |
H |
H |
F |
Cl |
TP30 |
H |
H |
CH3 |
H |
|
TP14 |
H |
H |
Cl |
Br |
TP31 |
H |
H |
OCH3 |
H |
|
TP15 |
H |
H |
F |
Br |
TP32 |
H |
Cl |
OCH3 |
Cl |
|
TP16 |
H |
Cl |
Cl |
Cl |
TP33 |
H |
H |
NHCOCH3 |
H |
|
TP17 |
H |
H |
Br |
Cl |
– |
– |
– |
– |
– |
Molecular docking of TPs
The molecular docking of TPs was performed utilizing three different proteins of SDM (PDB IDs: 3LD6, 5FSA, and 5TZ1) 18-20. The Molecular Operating Environment software (MOE) (2019.0102 version, Chemical Computing Group Inc., Canada) was employed for this study. The PDB files of proteins (PDB IDs: 3LD6, 5FSA, and 5TZ1) were downloaded from the Protein Databank website (RCSB.org). Each protein (3LD6, 5FSA, and 5TZ1) was uploaded in the software separately, purified by pressing the Quickpro button, and saved in the computer. Similarly, the Mole-Files of TPs, fluconazole, and ketoconazole were uploaded to the software, and their MDB files were prepared. The MDB files of TPs, fluconazole, and ketoconazole were docked with the purified proteins (3LD6, 5FSA, and 5TZ1) separately. The docking score (DS in kcal/mol) and the root mean square deviation (RMSD) were recorded (Table 2).
Prediction of the oral LD50 and toxicity class (TC) of TPs
The oral LD50 and TC of TPs, fluconazole and ketoconazole was predicted with ProTox II software 21. The Mole-Files of the compounds were imported into the software using the import button, the start button was pressed, and the data was recorded (Table 3).
Prediction of the pharmacokinetic parameters of TPs
The pharmacokinetic properties of TPs, fluconazole, and ketoconazole were predicted with the Swiss-ADME database 21,22. The Mole-Files of the compounds were imported into the software using the import button, the run button was pressed, and the data was recorded. The %absorption was calculated using a formula (%Absorption = 109 – (0.345 x TPSA)), wherein TPSA refers to topological polar surface area (Table 3).
Synthesis of TP18, TP22, TP27 and TP33
A mixture of 3-hydrazineyl-5-(methylthio)-1H-1,2,4-triazole (0.1 moles), 4-(2-hydroxy-4-methylphenyl)-4-oxobutanoic acid (0.1 moles), and ethanol (50 ml) was refluxed for 6 hours. A solid was precipitated during the refluxing, which was hot-filtered with a Whatmann filter paper and dried to get the solid. The solid was recrystallized with ethanol to get pure TP18. TP22, TP27, and TP33 were prepared by a similar method by replacing 4-(2-hydroxy-4-methylphenyl)-4-oxobutanoic acid with 4-(5-chloro-2-hydroxyphenyl)-4-oxobutanoic acid, 4-(4-aminophenyl)-4-oxobutanoic acid, and 4-(4-acetamidophenyl)-4-oxobutanoic acid, respectively (Scheme 1) (Table 4).
 |
Scheme 1: Synthesis of TP18, TP22, TP27, and TP33. |
Antifungal activity evaluation
This was performed by serial dilution methods described in the previous publications 8,12. In short, various dilutions of different concentrations (100, 50, 25, 12.5, 6.25, and 3.125 µg/ml) of TPs, fluconazole, and ketoconazole were prepared. The sterile DMSO was used as a solvent to prepare different dilutions of TPs, fluconazole, and ketoconazole and also served as a control. The agar medium was used to grow seven fungi in separate petri dishes, dilutions of different concentrations of TPs, fluconazole, and ketoconazole were added to the plates, and the minimum inhibitory concentrations (MICs) of TPs, fluconazole, and ketoconazole were identified (Table 5).
Statistical analysis
SPSS was used for the statistical analysis of the experimental data (version 20, Chicago, IL, USA). The results are considered statistically significant if the p-value (N = 3; Mean ± SD) is less than 0.05.
Results
The chemical structures of thirty-three TPs (TP1 to TP33) were designed (Figure 1) (Table 1). The DS and RMSD values of these compounds were recorded by molecular docking study employing three different proteins of SDM (PDB IDs: 3LD6, 5FSA, and 5TZ1) 18-20 (Table 2).
Table 2: DS of the designed TPs obtained by molecular docking
|
Compound |
DS in kcal/mol |
Compound |
DS in kcal/mol |
||||
|
3LD6 |
5FSA |
5TZ1 |
3LD6 |
5FSA |
5TZ1 |
||
|
Fluconazole |
-8.18 |
-8.79 |
-9.18 |
TP17 |
-6.46 |
-8.07 |
-7.57 |
|
Ketoconazole |
-8.16 |
-8.86 |
-9.06 |
TP18 |
-8.27 |
-9.07 |
-9.42 |
|
TP1 |
-6.81 |
-7.88 |
-7.79 |
TP19 |
-6.93 |
-7.34 |
-7.23 |
|
TP2 |
-7.05 |
-7.96 |
-7.17 |
TP20 |
-6.74 |
-6.97 |
-6.87 |
|
TP3 |
-7.26 |
-7.90 |
-7.68 |
TP21 |
-6.30 |
-7.42 |
-7.20 |
|
TP4 |
-6.72 |
-7.57 |
-8.31 |
TP22 |
-8.23 |
-8.93 |
-9.57 |
|
TP5 |
-7.02 |
-7.74 |
-8.00 |
TP23 |
-6.86 |
-7.58 |
-7.79 |
|
TP6 |
-6.78 |
-7.39 |
-7.52 |
TP24 |
-6.40 |
-7.82 |
-7.18 |
|
TP7 |
-6.32 |
-7.36 |
-7.91 |
TP25 |
-6.52 |
-7.75 |
-7.85 |
|
TP8 |
-6.86 |
-7.50 |
-7.10 |
TP26 |
-6.65 |
-7.32 |
-7.55 |
|
TP9 |
-6.83 |
-7.30 |
-7.84 |
TP27 |
-8.31 |
-9.12 |
-9.38 |
|
TP10 |
-6.26 |
-7.45 |
-7.61 |
TP28 |
-6.21 |
-7.33 |
-7.48 |
|
TP11 |
-6.16 |
-7.21 |
-7.11 |
TP29 |
-6.44 |
-7.22 |
-7.40 |
|
TP12 |
-6.16 |
-6.94 |
-7.43 |
TP30 |
-6.53 |
-7.61 |
-7.28 |
|
TP13 |
-6.29 |
-7.56 |
-7.13 |
TP31 |
-7.16 |
-7.95 |
-7.49 |
|
TP14 |
-6.59 |
-7.90 |
-7.61 |
TP32 |
-7.68 |
-7.39 |
-7.84 |
|
TP15 |
-6.36 |
-7.37 |
-7.49 |
TP33 |
-8.19 |
-8.98 |
-9.94 |
|
TP16 |
-6.65 |
-7.55 |
-7.62 |
– |
– |
– |
– |
In a docking experiment, the high negative value of the DS reflects the potency of the compound 21. The DS of TP18 (3LD6 = -8.27; 5FSA = -9.07; 5TZ1 = -9.42), TP22 (3LD6 = -8.23; 5FSA = -8.93; 5TZ1 = -9.57), TP27 (3LD6 = -8.31; 5FSA = -9.12; 5TZ1 = -9.38), and TP33 (3LD6 = -8.19; 5FSA = -8.98; 5TZ1 = -9.94) were better than the DS of fluconazole (3LD6 = -8.18; 5FSA = -8.79; 5TZ1 = -9.18) and ketoconazole (3LD6 = -8.16; 5FSA = -8.86; 5TZ1 = -9.06). This result implies high potency of TP18, TP22, TP27 and TP33 than fluconazole and ketoconazole. The interaction of fluconazole, ketoconazole, TP18, TP22, TP27, and TP 33 with 3LD6 protein is depicted in Figure 2a, Figure 2b, Figure 2c, Figure 2d, Figure 2e, and Figure 2f, respectively.
 |
Figure 2a: Interaction of fluconazole with 3LD6 protein of SDM. |
 |
Figure 2b: Interaction of ketoconazole with 3LD6 protein of SDM. |
 |
Figure 2c: Interaction of TP18 with 3LD6 protein of SDM. |
 |
Figure 2d: Interaction of TP22 with 3LD6 protein of SDM. |
 |
Figure 2e: Interaction of TP27 with 3LD6 protein of SDM |
 |
Figure 2f: Interaction of TP 33 with 3LD6 protein of SDM. |
Based on the DS of the compounds, the toxicity and pharmacokinetic parameters of TP18, TP22, TP27, and TP33 were predicted using the PtoTox II Web server 21 (Table 3), Swiss-ADME database 21,22 (Table 3), and the % absorption was also calculated utilizing the score of TPSA 21.
Table 3: Predicted toxicity and pharmacokinetic data of TPs
|
Compound |
ProTox II software |
Swiss-ADME data |
|||||||
|
predicted LD50 in mg/kg (Toxicity class) |
TPSA (Log Po/w) |
BBB permeant |
P-gp substrate |
CYP2C9 inhibitor |
CYP2D6 inhibitor |
CYP3A4 inhibitor |
Drug-likeness (Lipinski) |
Oral absorption |
|
|
Fluconazole |
1271 (4) |
81.65 (0.88) |
No |
Yes |
No |
No |
No |
Yes |
High |
|
Ketoconazole |
166 (3) |
69.06 (3.57) |
Yes |
No |
Yes |
Yes |
Yes |
Yes |
High |
|
TP18 |
500 (4) |
119.77 (1.76) |
No |
No |
No |
No |
No |
Yes |
High |
|
TP22 |
500 (4) |
119.77 (2.05) |
No |
No |
No |
No |
No |
Yes |
High |
|
TP27 |
1000 (4) |
125.56 (1.27) |
No |
Yes |
No |
No |
No |
Yes |
High |
|
TP33 |
1000 (4) |
128.64 (1.33) |
No |
Yes |
No |
No |
No |
Yes |
High |
A high value of LD50 and a low value of the toxicity class (TC) implies that a compound is relatively non-toxic compared to the standard drug 23. The LD50 and toxicity class (TC) of TP18 (500 mg/kg; TC 4), TP22 (500 mg/kg; TC 4), TP27 (1000 mg/kg; TC 4), and TP33 (1000 mg/kg; TC 4) was better than ketoconazole (166 mg/kg; TC 3). The TC of TP18, TP22, TP27, and TP33 was equal to fluconazole, but the LD50 of TPs was less than fluconazole (LD50 = 1271 mg/kg). All the compounds passed Lipinski’s drug-likeliness rule and demonstrated high oral absorption. None of the compounds displayed inhibitory activity against the metabolizing enzymes (CYP2C9, CYP2D6, and CYP3A4) and did not show the capability of crossing the blood-brain barrier (BBB). The bioavailability radar of TP18, TP22, TP27, and TP33 was also comparable to ketoconazole and fluconazole (Figure 3). In Figure 3, the red line represents the physicochemical characteristics affecting the compounds’ bioavailability, while the pink zone area denotes a region of acceptable bioavailability. The compound is bioavailable if the red line stays within the pink zone.
 |
Figure 3: Bioavailability radar of fluconazole, ketoconazole, TP18, TP22, TP27 and TP33. |
Based on the data mentioned above in silico studies, TP18, TP22, TP27, and TP33 were selected for their synthesis (Scheme 1) 16,17. The synthesized compounds’ spectral data (FTIR, 1H-NMR, 13C-NMR, and Mass) aligned to their designed chemical structure (Table 4). The synthesized compounds exhibited characteristic peaks for the carbonyl group of the pyridazinone ring, two methylene groups of the pyridazinone ring, the -NH- group of the triazole ring, and the substituents of the phenyl ring (-OH, -NH- and C=O groups) (Table 4).
Table 4: Characterization data of TP18, TP22, TP27 and TP33
|
Compound (MF; MW; M.P.; Rf value*; FTIR in KBr, ν in cm-1) |
1H-NMR (DMSO-d6, 500 MHz, δ in ppm) |
13C-NMR (DMSO-d6, 125 MHz, δ in ppm) |
Mass (m/z) |
|
TP18 (C14H15N5O2S; 317; 182-184oC; 0.77; 3350 (OH), 3233 (NH), 1712 (C=O)) |
2.20 (s, 3H, CH3), 2.41 (t, 2H, C4-methylene), 2.47 (s, 3H, CH3-S-), 2.90 (t, 2H, C5-methylene), 6.75 (s, 1H, Ar-H), 6.81 (d, 1H, Ar-H), 7.55 (d, 1H, Ar-H), 10.21 (s, IH, OH), 11.75 (s, IH, NH) |
13.4 (CH3-S-), 20.5 (CH3), 23.6 (C5-pyridazinone), 31.3 (C4-pyridazinone), 114.7, 118.3, 122.7, 126.1, 141.0, 145.4, 156.1, 158.1 (N-C-S), 160.1 (C-OH), 167.1 (C=O) |
317 (M+, 100%), 302, 210, 203, 196, 114, 107, 100, 97 |
|
TP22 (C13H12ClN5O2S; 337; 167-169oC; 0.73; 3352 (OH), 3234 (NH), 1713 (C=O)) |
2.41 (t, 2H, C4-methylene), 2.47 (s, 3H, CH3-S-), 2.91 (t, 2H, C5-methylene), 6.93 (d, 1H, Ar-H), 7.32 (d, 1H, Ar-H), 7.64 (s, 1H, Ar-H), 11.75 (s, IH, NH), 13.11 (s, IH, OH) |
13.4 (CH3-S-), 23.6 (C5-pyridazinone), 31.3 (C4-pyridazinone), 117.5, 119.1, 126.1, 129.5, 132.5, 145.4, 156.1, 158.1 (N-C-S), 159.5 (C-OH), 167.1 (C=O) |
337 (M+, 100%), 338 (M++1), 339 (M++2), 322, 223, 210, 196, 127, 114, 100, 97 |
|
TP27 (C13H14N6OS; 302; 173-175oC; 0,75; 3287 (NH2), 3233 (NH), 1712 (C=O)) |
2.42 (t, 2H, C4-methylene), 2.46 (s, 3H, CH3-S-), 2.90 (t, 2H, C5-methylene), 5.46 (s, 2H, NH2), 6.85 (d, 2H, Ar-H), 7.61 (d, 2H, Ar-H), 11.75 (s, 1H, NH) |
13.4 (CH3-S-), 23.3 (C5-pyridazinone), 31.3 (C4-pyridazinone), 113.2 (2C), 125.3, 129.1 (2C), 145.4, 149.6, 156.1, 158.1 (N-C-S), 167.1 (C=O) |
302.09 (M+, 100%), 287, 210, 196, 188, 114, 100, 97, 92 |
|
TP33 (C15H16N6O2S; 344; 179-181oC; 0.82; 3245 (NH), 3233 (NH), 1712, (C=O), 1705 (C=O)) |
2.04 (s, 3H, CH3-CO-), 2.41 (t, 2H, C4-methylene), 2.46 (s, 3H, CH3-S-), 2.90 (t, 2H, C5-methylene), 7.65 (d, 2H, Ar-H), 7.86 (d, 2H, Ar-H), 10.11 (s, 1H, -NH-CO-), 11.76 (s, 1H, -NH-) |
13.4 (CH3-S-), 23.1 (CH3-CO-), 23.3 (C5-pyridazinone), 31.3 (C4-pyridazinone), 120.6 (2C), 128.3 (2C), 131.1, 139.7, 145.4, 156.1, 158.1 (N-C-S), 167.1 (C=O), 167.6 (C=O, acetyl) |
344 (M+, 100%), 329, 301, 287, 230, 210, 196, 134, 114, 100, 97 |
*Rf values in a benzene and acetone mixture (8:2).
The in vitro antifungal activity of the synthesized compounds (TP18, TP22, TP27, and TP33) was carried out against seven fungi (Table 5) by serial dilution method 8,12.
Table 5: In vitro antifungal activity data of TP18, TP22, TP27 and TP33
|
Compound |
Zone of inhibition in mm (MIC in μg/ml)* |
||||||
|
C. albicans |
C. tropicalis |
A. fumigatus |
A. niger |
A. flavus |
P. citrinum |
M. purpureous |
|
|
Fluconazole |
25.04±0.23 (12.5) |
24.75±0.35 (12.5) |
22.22±0.07 (12.5) |
21.16±0.48 (12.5) |
18.18±0.14 (12.5) |
16.15±0.25 (12.5) |
25.31±0.44 (12.5) |
|
Ketoconazole |
24.12±0.45 (12.5) |
23.17±0.15 (12.5) |
20.06±0.32 (12.5) |
20.35±0.50 (12.5) |
20.45±0.33 (25) |
18.34±0.41 (25) |
24.22±0.24 (12.5) |
|
TP18 |
25.55±0.33 (6.25) |
24.62±0.11 (6.25) |
23.24±0.18 (6.25) |
22.22±0.27 (6.25) |
22.41±0.15 (12.5) |
19.98±0.24 (12.5) |
25.86±0.30 (6.25) |
|
TP22 |
28.83±0.16 (6.25) |
27.33±0.25 (6.25) |
24.82±0.36 (6.25) |
23.40±0.44 (6.25) |
23.55±0.50 (6.25) |
22.21±0.31 (6.25) |
27.37±0.16 (6.25) |
|
TP27 |
25.96±0.45 (12.5) |
25.79±0.28 (12.5) |
23.55±0.50 (12.5) |
22.62±0.35 (12.5) |
19.77±0.44 (12.5) |
18.88±0.30 (12.5) |
26.71±0.48 (12.5) |
|
TP33 |
25.55±0.15 (12.5) |
25.41±0.36 (12.5) |
22.98±0.49 (12.5) |
22.21±0.33 (12.5) |
19.50±0.28 (12.5) |
18.80±0.46 (12.5) |
26.34±0.14 (12.5) |
|
Control |
0.0±0.0 |
0.0±0.0 |
0.0±0.0 |
0.0±0.0 |
0.0±0.0 |
0.0±0.0 |
0.0±0.0 |
*p < 0.05.
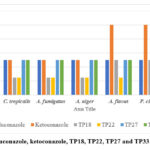 |
Figure 4: MIC of fluconazole, ketoconazole, TP18, TP22, TP27 and TP33 against tested fungi |
The antifungal activity data (Table 5 and Figure 4) implies that TP18, TP22, TP27, and TP33 are better antifungal agents than fluconazole and ketoconazole. These findings concur with the DS of the compounds (Table 1).
Discussion
This work relates to the discovery of safer and more potent TPs (Figure 1) (Table 1) as an inhibitor of SDM. Among the thirty-three designed TPs, four TPs (TP18, TP22, TP27, and TP33) displayed better DS (an indicator of the compound’s potency) than fluconazole and ketoconazole. This observation implies that TP18, TP22, TP27, and TP33 are more potent inhibitors than fluconazole and ketoconazole. It was observed that fluconazole, ketoconazole, TP18, TP22, TP27 and TP33 interact with many common amino acids (Arg382, Arg446, Arg448, Cys449, Gly307, Gly312, Gly443, Gly451, His447, Lys156, Ser316, Thr135, Thr315, Thr319, Tyr107, Tyr131, and Tyr145) of 3LD6 protein of SDM enzyme (Figure 2a, Figure 2b, Figure 2c, Figure 2d, Figure 2e, and Figure 2f, respectively). This observation indicates that fluconazole, ketoconazole, TP18, TP22, TP27, and TP33 share the same mechanism of action and inhibit SDM enzyme by binding at a common site of SDM. The in silico studies of TP18, TP22, TP27, and TP33 also revealed their non-toxic behavior (Table 3), promising pharmacokinetic properties (Table 3), and acceptable bioavailability (Figure 3). Accordingly, TP18, TP22, TP27, and TP33 were synthesized by reacting 3-hydrazineyl-5-(methylthio)-1H-1,2,4-triazole and 4-Oxobutanoic acid derivatives (Scheme 1). The spectral data (FTIR, 1H-NMR, 13C-NMR and Mass) of TP18, TP22, TP27, and TP33 (Table 4). The antifungal activity data implies that TP18, TP22, TP27, and TP33 were better antifungal agents than fluconazole and ketoconazole against all seven tested fungi. These findings concurred with the DS of TP18, TP22, TP27, and TP33. The structure-activity relationship of TP18, TP22, TP27, and TP33 indicates that the presence of at least one hydrophilic group like OH (TP18 and TP22), NH2 (TP27), and -NH-CO-CH3 (TP33) provide good SDM inhibitors. A second substituent, like methyl group (TP18) and chloro group (TP22), may synergize the SDM inhibitory effects. Accordingly, the incorporation of a methyl group or a chloro group in TP27 and TP33 may also synergize their SDM inhibition property.
Many structural changes are possible in TPs for further research. Azole is a general term for a five-membered heterocyclic ring containing at least one nitrogen and other hetero atoms (S, O, and N). The designed TPs contain a triazole ring, a pyridazinone ring, and a phenyl ring (Figure 1). Replacing the triazole ring with other azole rings (imidazole, pyrazole, tetrazole, pentazole, oxazole, thiazole, and isoxazole) can get better SDM inhibitors. The same idea applies to replacing pyridazinone and phenyl rings with their known bio-isosteres. Medicinal thiols (-SH group containing molecules) are important in the literature. The designed TPs contain the CH3-S- group, which may be replaced with the thiol group. The LD50 values, toxicity profile, pharmacokinetic parameters, oral absorption, bioavailability parameter, metabolizing enzyme inhibitory potential, and drug-likeliness characteristics of TP18, TP22, TP27, and TP33 have been appreciable (Table 3) (Figure 3) in comparison to fluconazole and ketoconazole. However, any structural change in TP18, TP22, TP27, and TP33 or other TPs may also alter these properties. Accordingly, this aspect needs attention while designing analogs of the designed TPs. TP18, TP22, TP27, and TP33 demonstrated appreciable in vitro antifungal activity against seven fungi (Table 5). The antifungal activity data of these compounds looks good but does not guarantee their broad-spectrum properties. Similarly, the toxicity profile and pharmacokinetic parameters of TP18, TP22, TP27, and TP33 are based on their in silico studies. Accordingly, there is a need to assess the antifungal activity of TP18, TP22, TP27, and TP33 against various pathogenic fungi, including drug-resistant fungi, and to carry out various in vitro and in vivo toxicity assays. Azole derivatives also possess antibacterial activity 8. Therefore, the antibacterial activity evaluation of the designed TPs (Figure 1) is also recommended.
Conclusion
This work relates to the discovery of safer and more potent TPs as an inhibitor of SDM. Four compounds (TP18, TP22, TP27, and TP33) have been recognized as safe, effective, and potent inhibitors of SDM, holding appreciable in silico study-based toxicity profiles and pharmacokinetic properties. Even so, their broad-spectrum antifungal activity and in vivo toxicity profiles are yet to be established. The possibility of many structural modifications in these compounds makes them an excellent new chemical template for developing new and better broad-spectrum antifungal agents to combat AMR. Accordingly, additional study is advised on TP18, TP22, TP27, and TP33 to confirm these expectations.
Acknowledgment
The authors gratefully acknowledge the approval and the support of this research study by grant no. SCIA-2023-12-2264 from the Deanship of Scientific Research at Northern Border University, Arar, K.S.A.
Conflict of Interest
No conflict of interest is associated with this work.
References
- Tang, K.W.K.; Millar, B.C.; Moore, J.E. Br. J. Biomed. Sci., 2023, 80, 11387.
CrossRef - Carmo, A.; Rocha, M.; Pereirinha, P.; Tomé, R.; Costa, E. Antibiotics (Basel)., 2023, 12(5), 884.
CrossRef - Moyo, P.; Moyo, E.; Mangoya, D.; Mhango, M.; Mashe, T.; Imran, M.; Dzinamarira, T. J. Infect. Pub. Health 2023, 16(4), 632-639.
CrossRef - Cortés, J.C.G.; Curto, M.Á.; Carvalho, V.S.D.; Pérez, P.; Ribas, J.C. Biotechnol. Adv., 2019, 37(6), 107352.
CrossRef - Zhen, C.; Lu, H.; Jiang, Y. Front. Microbiol., 2022, 13, 911322.
CrossRef - Singh, A.; Singh, K.; Sharma, A.; Kaur, K.; Chadha, R.; Bedi, P.M.S. Chem. Biol. Drug Des., 2023, 102(3), 606-639.
CrossRef - Monk, B.C.; Keniya, M.V. J. Fungi (Basel)., 2021, 7(2), 67.
CrossRef - Imran, M.; Bawadekji, A.; Alotaibi, N. Trop. J. Pharm. Res., 2020, 19(2), 377-382.
CrossRef - Akhtar, W.; Shaquiquzzaman, M.; Akhter, M.; Verma, G.; Khan, M.F.; Alam, M.M. Eur. J. Med. Chem., 2016, 123, 256-281.
CrossRef - Rozada, A.M.F.; Rodrigues-Vendramini, F.A.V.; Gonçalves, D.S.; Rosa, F.A.; Basso, E.A.; Seixas, F.A.V.; Kioshima, É.S.; Gauze, G.F. Bioorg. Med. Chem. Lett., 2020, 30(14), 127244.
CrossRef - Imran, M.; Asif, M. Russ. J. Bioorg. Chem., 2020, 46, 726-744.
CrossRef - Abida; Diwan, A.; Thabet, H.K.; Imran, M. Trop. J. Pharm. Res., 2019, 18(12), 2633-2641.
- Saini, M.; Mehta, D.K.; Das, R.; Saini, G. Mini Rev. Med. Chem. 2016, 16(12), 996-1012.
CrossRef - Sukuroglu, M.; Onkol, T.; Onurdağ, F.K.; Akalin, G.; Sahin, M.F. Z. Naturforsch. C. 2012, 67(5-6), 257-65.
CrossRef - Zhou, G.; Ting, P.C.; Aslanian, R.; Cao, J.; Kim, D.W.; Kuang, R.; Lee, J.F.; Schwerdt, J.; Wu, H.; Herr, R.J.; Zych, A.J.; Yang, J.; Lam, S.; Wainhaus, S.; Black, T.A.; McNicholas, P.M.; Xu, Y.; Walker, S.S. Bioorg. Med. Chem. Lett. 2011, 21(10), 2890-2893.
CrossRef - James, F.D.; Joseph, K.P. United States Patent Number US3331840A., 1967. https://patents.google.com/patent/US3331840A/en?oq=US3331840A (Accessed on August 26, 2023)
- Jojima, T.; Takahi, Y. United States Patent Number US4052395A., 1977. https://patents.google.com/patent/US4052395A/en?oq=US4052395A (Accessed on August 26, 2023).
- Sanaullah, A.F.M.; Devi, P.; Hossain, T.; Sultan, S.B.; Badhon, M.M.U.; Hossain, M.E.; Uddin, J.; Patwary, M.A.M.; Kazi, M.; Matin, M.M. Molecules., 2023, 28(3), 986.
CrossRef - Khichi, A.; Jakhar, R.; Dahiya, S.; Arya, J.; Dangi, M.; Chhillar, A.K. J. Biomol. Struct. Dyn., 2023, 1-14.
CrossRef - Uppar, V.; Chandrashekharappa, S.; Shivamallu, C.P.S.; Kollur, S.P.; Ortega-Castro, J.; Frau, J.; Flores-Holguín, N.; Basarikatti, A.I.; Chougala, M.; Mohan, M.M.; Banuprakash, G.; Venugopala, K.N.; Nandeshwarappa, B.P.; Veerapur, R.; Al-Kheraif, A.A.; Elgorban, A.M.; Syed, A.; Mudnakudu-Nagaraju, K.K.; Padmashali, B.; Glossman-Mitnik, D. Molecules., 2021, 26(9), 2722.
CrossRef - Imran, M.; Mohd, A.A.; Nayeem, N.; Alaqel, S.I. Trop. J. Pharm, Res., 2023, 22(6), 1263-1269.
- Daina, A.; Michielin, O.; Zoete, V. Sci. Rep., 2017, 7, 42717.
CrossRef - Gadaleta, D.; Vuković, K.; Toma, C.; Lavado, G.J.; Karmaus, A.L.; Mansouri, K.; Kleinstreuer, N.C.; Benfenati, E.; Roncaglioni, A. J. Cheminform., 2019, 11(1), 58.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









