Synthesis of CeO2-GO Nano Composite and its Impact on SOD1 Protein Through Computation Study: Molecular Docking
S. Chitrarasu1 , A. Selvam2
, A. Selvam2 , M. Yogapriya1*
, M. Yogapriya1* , K. Boopathi3
, K. Boopathi3 and K.Selvapriya4
and K.Selvapriya4
1Department of Chemistry, Kalaignar Karunanidhi Government Arts College, Tiruvannamalai, (Tamilnadu), India.
2Department of Chemistry, Panimalar Enginering College,Chennai,Tamilnadu,India.
3Department of Chemistry, Pondicherry University, Kalapet, (Pondicherry), India.
4Department of Bioinformatics, Pondicherry University, Kalapet, (Pondicherry), India.
Corresponding Author E-mail: yogapriyam@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/390523
Article Received on : 19 Aug 2023
Article Accepted on : 06 Oct 2023
Article Published : 17 Oct 2023
Reviewed by: Dr. Hamid Reza Ghorbani
Second Review by: Dr. Amit Sharma
Final Approval by: Dr.Mohammad Tawkir Sheikh
Motor neuron disease (MND) has become the most widespread neurodegenerative disease, like ALS, AD, and PD, affecting millions of human beings worldwide. Among these, ALS disease plays a major role in MND. The metal oxides have an essential role in neurodegenerative diseases. Cerium oxide nanoparticles have received a lot of attention in recent years as a potential future remedy for treating a variety of problems due to their redox activity, free radical scavenging capabilities, biofilm suppression, and other features. We are using molecular docking experiments to better understand the interaction of transition metal oxides (CeO2) with mutated and Non-mutated SOD1 proteins. Through the use of molecular docking studies, the structural relationship between amino acids, binding energy, and ligand efficiency was investigated. Auto-dock analysis also reveals that the CeO2 nanoparticle has significant binding energy. In this work, we synthesize a Cerium oxide/GO composite and examine its impact on mutant and non-mutant SOD1 proteins, as well as antioxidant assays (SOD3), anti-microbial activity, and CV analysis.
KEYWORDS:Cerium oxide/GO nanocomposite; CV; Molecular docking; superoxide dismutase ( SOD1)
Download this article as:| Copy the following to cite this article: Chitrarasu S, Selvam A, Yogapriya M, Boopathi K, Selvapriya K. Synthesis of CeO2-GO Nano Composite and its Impact on SOD1 Protein Through Computation Study: Molecular Docking. Orient J Chem 2023;39(5). |
| Copy the following to cite this URL: Chitrarasu S, Selvam A, Yogapriya M, Boopathi K, Selvapriya K. Synthesis of CeO2-GO Nano Composite and its Impact on SOD1 Protein Through Computation Study: Molecular Docking. Orient J Chem 2023;39(5). Available from: https://bit.ly/3rVPtNo |
Introduction
Nanoparticle development has branched out into a wide spectrum of clinical uses in the last few decades. Nanoparticles have been intended to overcome the limits of free treatments and to transportation biological barriers that are varied among different patients and disorders. Precision treatments, which use individualized interventions to improve therapeutic efficacy, have also helped to overcome patient heterogeneity. However, the research of nanoparticles remains focused on improving delivery systems with a one-size-fits-all approach. Nanoparticles can be manufactured and changed technologically to carry components for medical imaging, cancer treatment, or medication release. Because of the conjugate’s versatility, nanomaterials have been widely employed in diagnostics and therapy. The effects of nanoparticles on cells are yet unknown, in addition to the newly developed uses of nanomaterial in biological systems. Numerous studies on the effects of nanomaterial’s on biological systems show the need for a completely novel area of study specialized in Nano toxicity 1 . CeO2 nanoparticles have the potential to be developed as a treatment for oxidative stress diseases. Furthermore, in accordance with the conditions, Cerium may change oxidation states (Ce3+ or Ce4+) and bind oxygen reversibly. Cerium oxide have been studied in biological systems because to their properties and have determined antioxidant effects in numerous disease models. Nanoceria has significant antibacterial action against both Gram-positive and Gram-negative bacteria via the production of reactive oxygen species (ROS)2. The antioxidant activity of cerium oxide nanoparticles is reversible. It has a wide range of possible uses in pharmacology. This neutralizing function is similar to that of metabolic enzymes such as superoxide dismutase (SOD) and catalase, which are ROS scavengers. As a result, cerium oxide nanoparticles can protect cells from oxidative damage in the environment 3. Nano ceria demonstrates both catalysis and SOD-mimetic action4-7. Cerium nanoparticles having bactericidal action might be employed in the production of medical and food processing equipment, among other sectors. The biological activity of highly reactive cerium oxide nanoparticles against Gram (+) and Gram (-) bacteria is good. 8 The development of artificial intelligence and machine learning (AI and ML) has made it possible to find lead compounds by using computerized database searches that target specific proteins 9. Additionally, the early stages of medication development research concentrate on a particular disease’s root cause. Modern pharmacological advancements have made it possible to better understand the cellular and molecular causes of illnesses 10. Because of this, the majority of pharmaceutical companies and academic research projects start by choosing an appropriate target inside the body, then thoroughly examining the signaling pathways and developing a drug to interactions with proteins 11. Investigating the target’s structural and functional characteristics as well as how it interacts with compounds that have therapeutic properties 12, predicting the binding of microscopic drug-like substances to target proteins was the main objective of molecular docking 13. Protein malfunction is a common cause of disease, and medications work by either blocking or activating the target proteins 14. Traditional methods for generating drug development leads frequently entail testing a large number of intriguing compounds against a known disease target protein and hoping to detect a binding association 15. In a way similar to investigational high-throughput screening, docking may be used to digitally screen new compounds, and it can also be utilized to offer atomistic-level information to support structure-based drug development 16.
Materials, Methods and Characterizations.
Materials
Graphite powder, nitrate soda (NaNO3), concentrated sulfuric acid (conc. H2SO4), potassium permanganate (KMnO4), hydrogen peroxide (H2O2), Ammonium hydroxide(NH4OH), cerous nitrate hexahydrate Ce(NO3)36H2O, hydrazine (N2H4) were bought from Proxor, India.
Preparation of GO
The GO was set up using the modified Hummers method. 10 g of powdered graphite and 5 g of NaNO3 were added slowly while stirring to a 240 ml solution of ice-cold H2SO4 in water. 32 g of KMnO4 were added to this solution within two hours. 250 cc of pure water was carefully added to the reactants, and then the reactants were heated to below 40 °C and stirred for 90 minutes. The reaction mixture temperature was monitored and held at 90 degrees Celsius for 60 minutes. 800 ml of distilled water and 23 ml of a 25% aqueous solution of H2O2 were combined to complete the reaction. The resulting solution was repeatedly washed with distilled water until the filtrate’s pH reached neutral. The resulting solution was subjected to 20 minutes of sonication and 70°C oven drying 17-19.
Synthesis of the CeO2/GO nanocomposite
In 30 ml of distilled H2O, one millimol (0.4342 g) of Ce(NO3)3.6H2O was dissolved, and 2 ml of 32% NH4OH (aqueous) was dropped into the solution using sonication. The entire procedure took 66 minutes. Then add 60 mL of 0.5 mg NaOH aqueous. 1 gram of GO suspension was added and heated to 90 degrees Celsius. After adding 3 mL of N2H4, the solution was refluxed for 1 hour. The product was continually washed with ethanol and deionized water to create the cerium oxide-graphene oxide nanocomposite before being dried at 60 °C for 24 hours20,21.
Characterizations
The X-Ray Diffractometer from PANalytical was able to record X-Ray Diffraction patterns between 2 = 10 ° and 80 °. HRTEM using a JEOL JEM 2100, FTIR, and biological examinations were carried out by Acme ProGen Biotech (India) Private Limited, a NABL-accredited company. Docking studies were done with an authorized tool called MGL Tools, which has the software AUTODOCK (version 1.5.7) and AUTODOCK VINA (v1.2.x).
Results and Discussion
XRD analysis
 |
Figure 1: Shows XRD pattern of CeO2-GO. |
The XRD analysis of CeO2-graphene oxide nanocomposites The sample has XRD peaks at 28.59, 33.14, 47.44, 56.3, 58.95, and 69.4, as shown in the figure. The six peaks correspond to the cubic fluorite CeO2 (JCPDS 34-0394) crystal planes (1 1 1), (2 0 0), (2 2 0), (3 1 1), (2 2 2), and (4 0 0). The findings show that the nanocomposite is made up of CeO2 and graphene oxide, with no additional chemicals synthesized throughout the process22,23.
TEM analysis
|
|
Figure 2: illustrate HRTEM pattern of CeO2-GO. |
The Morphological analysis of GO@CeO2 composites is provided by the HRTEM investigation, which also establishes the size and shape of CeO2 nanoparticles. A high number of CeO2 nanoparticles are regularly dispersed on the surface of the transparent GO sheet in Figure 2c, which also shows distinct profiles for the GO sheet. The structures on graphene oxide foils or sheets, which provided the active centers for CeO2 deposition, may be used to explain it. CeO2 nanoparticles had a diameter of around 10–20 nm, as seen in Figure 2e.
EDX
 |
Figure 3: illustrate EDX pattern of CeO2-GO. |
Moreover, the CeO2/GO nanocomposite was found using EDX. The emission of substantial oxygen and cerium peaks, as well as a considerable carbon peak, are used to support the synthesis of CeO2 nanostructures and GO.
FTIR Spectrum
 |
Figure 4: illustrate FTIR pattern of CeO2-GO |
The vibrational stretching for OH group at 3050–3650 cm-1, C=O at 1700–1800 cm-1, COO– at 1600–1750 cm-1, sp2-hybridised Carbon (in-plane stretching) at 1450–1600 cm-1 are visible in the layer GO FTIR spectrum shown in Fig 4. at room temperature. sp2-hybridized Carbon is found between 1500 and 1670 cm-1. The existence of ceria in the solid was confirmed by the appearance of a broad band in all spectra between 400 and 630 cm-1, which may be recognized to the Ce-O’s stretching mode of tension and corresponds to the active F1U mode in the IR for the fluorite structure 24.
Docking studies
Proteins selection and preparation for docking studies.
Structure of Human Cu,Zn Superoxide Dismutase
The target protein was SOD1 precursor protein for ALS disease (PDB ID: 1PU0). The Protein Data Bank of the Research Collaboratory for Structural Bioinformatics (RCSB) (http://www.rcsb.org/pdb) was used to determine its structure. Its 3D structure has a resolution of 5 Å, chains of A,B,C,D,E , Sequence Length 153,homo sapiens ,non- mutated protein25.
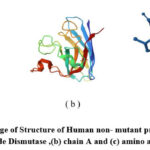 |
Figure 5: illustrate image of Structure of Human non- mutant protein SOD1 (a) Cu,Zn Superoxide Dismutase ,(b) chain A and (c) amino acids only |
Structure of Human Superoxide Dismutase (A4v Mutant) in C2 Space Group.
The target protein was SOD1 precursor protein for ALS disease (PDB ID: 6SPA). The Protein Data Bank of the Research Collaboratory for Structural Bioinformatics (RCSB) (http://www.rcsb.org/pdb) was used to determine its structure. Its 3D structure has a resolution of 5 Å, chains of A,B,C,D,E , Sequence Length 153,homo sapiens , mutant protein26.
 |
Figure 6: illustrate image of Structure of Human mutated protein SOD1 (a) Cu,Zn Superoxide Dismutase ,(b) chain A and (c) amino acids only |
Non-Mutant SOD1 Protein – Ligand interaction
 |
Figure 7: illustrate (a) image of interaction between protein and ligand and (b) binding sites of ligand in protein |
Mutant SOD1 Protein – Ligand interaction
 |
Figure 8: illustrate (a) image of interaction between protein and ligand and (b) binding sites of ligand in protein. |
Docking tools
The Protein Data Bank database (PDB) with IDs 1PU0 and 6SPA was used to download the protein structure. CeO2‘s small molecule structure was found in the PubChem database. Studies on interactions were carried out independently for the protein structure and the four small molecules. Docking investigations were carried out using the software Autodock (version 1.5.7) and Autodock Vina (v1.2.x) from MGL Tools, an authorized tool. Please reference AutoDock Vina in your work by using the following citation information. AutoDock Vina: Enhancing the Efficiency and Reliability of Docking with an Unique Scoring Function, Rapid Optimization, and Multithreading,” by O. Trott and A. J. Olson, Journal of Computational Chemistry, 31 (2010), 455–461. For further details, visit http://vina.scripps.edu. The search space volume is more than 27000 Angstrom3; warning (see FAQ). It was done to read the input, set up the scoring function, examine the binding site, search using a random seed of 915023360, and refine the findings27.
Autodock Vina
The protein structure was downloaded and viewed with the visualization software PYMOL for the structural formation, number of chains, and missing atoms. Protein was stored in PDB format for the further preparation process. Small molecules are downloaded in SDF format from PubChem and changed into PDB format with the help of PYMOL for future use. Before docking, we have to prepare the protein and ligand and save them in.pdbqt format. To do that, we need an auto-dock tool. In protein preparation, we remove water from the protein, add hydrogen, add charges, and save in.pdbqt format. Same way, prepare ligands with additions for torsions and save in.pdbqt format. Now grid formation, for blind docking all residues, is covered in a grid box, and the parameters are saved in.txt format for the configuration file. Before starting the docking process, we need a configuration file that contains an axis range and centre for the grid and is saved in.txt format 28. Auto dock Vina runs docking in a command prompt, opens the command prompt input path details, and runs docking; the result will be in output.pdbqt format. Results analyzed in the auto-dock and interactions were studied. After docking, we will get log.txt and output.pdbqt, which contain all the processes and parameters of our docking process29.
Protein- ligand -Interaction Parameters
 |
Figure 9: illustrate 1PUO with CeO2 interaction parameters |
 |
Figure 10: illustrate 6SPA with CeO2 interaction parameters. |
Docking results
Table 1 represents docking results in detail
Table.-1: illustrate docking results
|
Metal Oxide |
Protein ID |
Chain |
Residues |
Hydrogen Bonding Interaction |
Amino acid interactions |
Docking Score |
|
CeO2 |
Mutated 6SPA |
A |
153 |
02 |
THR116, ASP52 |
-2.6 |
|
Non Mutated 1PU0 |
A |
153 |
03 |
ASN139,THR137, ALA140,GLY141 |
-2.8 |
Protein with Cerium Oxide Confirmation Information
The interaction between the ligand CeO2 and protein 6SPA exhibits B.E. = -2.6 kcal/mol, and it creates two hydrogen bonds with amino acid residues including ASP52 and THR116.
The interaction between the ligand CeO2 and protein 1PU0 exhibits B.E. = -2.8 kcal/mol, and it creates three hydrogen bonds with amino acid residues including ASN139, THR137, ALA140, and GLY141.
to determine the mutant 6SPA chain A residues ASP52, and THR116 metal binding site for CeO2 as well as the binding energy (-2.6 kcal/mol). It was shown that CeO2 forms hydrogen bonds with charged and polarized residues like ASP52 and THR116 through interaction. CeO2 nanoparticle stability and nucleation formation are facilitated by a variety of contact forces, including the generation of hydrogen bonds between CeO2 and polar residues and the electrostatic force of attraction between CeO2 and charged residues in the binding sites of mutant 6SPA protein. CeO2 interactions between non-mutant 1PU0 chain A residues and GLY141, ASN139, ALA140, and THR137 result in interactions with the binding energy (-2.8 kcal/mol). CeO2 was shown to interact with both polar and charged residues such ASP139, GLY141and THR137.
Of all potential types—van der Waals, hydrophobic, solvation, torsional, and electrostatic—the electrostatic potential contributes the greatest to the overall binding energy. Similar interactions occur when the tiny molecule cerium oxide interacts with proteins and closely bonds to the amino acids aspartic acid, threonine, asparagine, alanine, and glycine.
Antioxidant Assay SOD (SOD 3)
 |
Figure 11: (a) image shows the inhibition % of SOD3 Assay and ( b) STD SOD3 Assay. |
Preparation of Standard SOD solution.
As we followed the procedure of Method for Clinical Assay of SuperoxideDismutas, Yi Sun et al.to measure SOD activity, Activity assays are capable of measuring SOD activity 30. In the Method for Clinical Assay of Superoxide Dismutas technique, nitroblue tetrazolium (NBT) reduction is employed as an indication of O2-production and xanthine-xanthine oxidase (XO) as the source of O2-. Since SOD and NBT compete for O2-, the percentage inhibition of NBT reduction serves as a gauge of the level of SOD in the environment. To eliminate the H2O2 that SOD produces, catalase is added. Units per milligramme of protein are used to measure the specific activity of enzymes. It is crucial to add the reagents into the test solution in the correct sequence. The initial protein concentration of the sample should be between 10 and 100 g. There is a linear relationship between sample concentration and inhibition percentage 31.
Determination of antibacterial activity by well-diffusion method
Inoculums Preparation
At minimum 3 to 5 well-isolated colonies of the similar morphological form are chosen from an agar plate culture. By lightly touching the top of every colony with a loop and transferring the development into a tube with 3 to 4 cc of a appropriate broth medium, such as enriched soy broth. 2. The broth culture is incubated at 35°C for approximately 3 to 6 hours, or until the turbidity of the 0.5 McFarland standard is reached or exceeded. Note: DMS serves as the negative control while the antibiotic tetracycline serves as the positive control. Both Gram (-) (E. coli) and Gram (+) (S. aureus) bacteria were resistant to the synthetic cerium oxide@GO nanocomposite’s antibacterial effects. According to our findings, compared to Gram-negative bacteria (E. coli), nanocomposite were more effective against Gram-positive bacteria (S. aureus). In this situation, cerium oxide@GO nanocomposite demonstrated the appropriate degree of bactericidal activity32-36.
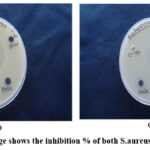 |
Figure 12: (a) image shows the inhibition % of both S.aureus and E.coli bacteria |
Cyclic voltammogram analysis
 |
Figure 13: image shows the Cyclic voltammogram of CeO2/GO nano composite at different scan rate. |
In cyclic voltammogram , The current Vs. potential characterizations of the synthesized Cerium oxide/GO Nano composite were measured. The reference electrode was stranded Ag/AgCl, the counter electrode was Pt. wire, and the working electrode was GCE. For all measurements, an aqueous solution of 1 M Na2So4 served as the electrolyte37.
Conclusion
The structural characteristics of the CeO2/GO nanocomposite, which had nanoparticle sizes ranging from 10 to 20 nm, were well generated in an environmentally benign manner. The sample concentration and inhibition percentage in the antioxidant test (SOD3) are correlated linearly. Higher inhibition percentages increase with higher sample concentrations. Cerium oxide@GO nanocomposite showed a satisfactory level of bactericidal activity. According to our conclusions, as related to Gram (-) bacteria (E. coli), nanocomposite was most efficient against Gram (+) bacteria (S. aureus). Docking studies were done for the SOD1 proteins with mutated 6SPA and non-mutated 1PU0 proteins with Cerium Oxide/GO. Docking results show there was a strong non-bonding interaction between CeO2 with both mutant and non-mutant proteins. GO acts as a better platform for delivering the CeO2 nanoparticles to the cell membrane. It does not affect that Protein environment. Docking results also reveal that GO has no docking score with the SOD1 protein; based on the results, GO solely serves as a carrier for metal oxide. With mutant and non-mutant SOD1 proteins, cerium oxide had docking scores of -2.6 and -2.8, respectively. Cerium oxide, a tiny molecule, interacts with proteins and has intimate nonbonding interactions with the amino acids threonine, aspartic acid, asparagine, alanine, and glycine. According to auto-dock investigations, CeO2 nanoparticles have a high docking score. CeO2 nanoparticles bind to the active sites of the SOD1 protein based on binding energy values. SOD1 protein aggregates less, more harmful superoxide anions are converted to less harmful oxygen molecules, and peroxide is produced. To prevent cell damage, this procedure lowers oxidative stress in the cell.
Acknowledgment
The authors acknowledge Kalainger Karunanidhi College of Arts and Science, Tiruvannamalai for helping in doing this research work.
Conflict of Interest
The authors declare no conflict of interest
References
- Mitchell, M.J.; Billingsle, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Nature reviews drug discovery. 2021, 20, 101-24.
CrossRef - Thakur, N.; Manna, P.; Das, J. Journal of nanobiotechnology. 2019, 17, 1-27.
CrossRef - Mehta, A.; Scammon, B.; Shrake, K.; Bredikhin, M.; Gil, D.; Shekunova, T.; Baranchikov, A.; Ivanov, V.; Reukov, V. Materials Science and Engineering: C. 2020 ,114,111003.
CrossRef - Estevez, A.Y.; Pritchard, S.; Harper, K.; Aston, J.W.; Lynch, A.; Lucky, JJ.; Ludington, JS.; Chatani, P.; Mosenthal, W.P.; Leiter, J.C.; Andreescu, S. Free Radical Biology and Medicine. 2011, 51, 1155-63.
CrossRef - Heckman, K.L.; DeCoteau, W.; Estevez, A.; Reed, K.J .;Costanzo, W.; Sanford, D.; Leiter, J.C.; Clauss, J.; Knapp, K.; Gomez, C.; Mullen, P. ACS nano. 2013 ,7,10582-96.
CrossRef - Hirst, S.M.; Karakoti, A.S.; Tyler, R.D.; Sriranganathan, N.; Seal, S.; Reilly, C.M. Small. 2009,5,2848-56.
CrossRef - Pirmohamed, T.; Dowding, J.M.; Singh, S.; Wasserman, B.; Heckert, E.; Karakoti, A.S.; King, J.E.; Seal, S.; Self, W.T. Chemical communications. 2010,46,2736-8.
CrossRef - Reshma, P.; Ashwini, K. J Nanomater Mol Nanotechnol . 2017,4,1-4.
- Mateev, E.; Georgieva, M.;Zlatkov, A. Biointerface Res. App. Chem.2022,13,159
CrossRef - Altayb, H.N.; Fludarabine. Pharmaceuticals. 2022 ,15,1129
CrossRef - SAHA, S.; JOSHI, B.C.; JUYAL, V.; SAH, A.N. Physical Chemistry Research. 2023 ,11,801-23
- Halder, D.;Das, S.; Aiswarya, R.; Jeyaprakash, R.S. RSC advances. 2022,12,21452-67
CrossRef - Lin, Y.F.; Cheng, C.W.; Shih, C.S.; Hwang, J.K.; Yu, C.S.; Lu, C.H. Journal of chemical information and modeling. 2016 ,56,2287-91.
CrossRef - Al-Madhagi, H.A. Digital Chinese Medicine. 2023,6,67-72.
CrossRef - Amer, H.H.;, Eldrehmy, E.H.; Abdel-Hafez, S.M.;Alghamdi, Y.S.; Hassan, M.Y.;Alotaibi, S.H. Scientific reports. 2021,11,17953.
CrossRef - Ainsley, J.; Lodola, A.; Mulholland, A.J.; Christov, C.Z.; Karabencheva-Christova, T.G. Advances in protein chemistry and structural biology. 2018, 113, 1-32.
CrossRef - Amba Sankar, K.N.; Sathish Kumar, C.; Mohanta, K. Materials Today: Proceedings. 2019,18,759-64.
CrossRef - Amba Sankar, K.N.; Nandakumar, P. Rasayan J. Chem. 2021,14,2196-201.
CrossRef - Nandakumar, P.; Amba Sankar, K.N.; Ganesh, A.S.; Anandh, B.A.; Deepa, R. Oriental Journal of Chemistry. 2022,38,604.
CrossRef - Hamid Reza Ghorbani, Chemical Engineering Communications, 2016,202(11),1463-1467, DOI: 10.1080/00986445.2014.950732
CrossRef - Hamid Reza Ghorbani, Orient. J. Chem., Vol. 30(2), 803-806 (2014), http://dx.doi.org/10.13005/ojc/300254
CrossRef - Sagadevan.S.;Johan,M.R.;Lett,J.A. Appl.Phys.A, 2019,125,315,https://doi.org/10.1007/s00339-019-2625-6.
CrossRef - Yi Wang.; Chun Xian Guo.; Jiehua Liu.;Tao Chen.; Hongbin Yanga .;Chang Ming Li. : Dalton Trans., 2011, 40, 6388, DOI: 10.1039/c1dt10397k
CrossRef - Amin Shiralizadeh Dezfuli.; Mohammad Reza Ganjali.; Hamid Reza Naderi .; Parviz Norouziab. RSC Adv., 2015, 00, 1-3DOI: 10.1039/C5RA02957K.
CrossRef - Liu, Yang.; Yang Cao.,Computational Drug Discovery and Design. 2023. 113-125.
CrossRef - Liu, Y.;Yang, X.;Gan, J.; Chen, S.; Xiao, Z. X.; Cao, Y. CB-Dock2, Nucleic acids research,2022, 50 ,159-164
CrossRef - O. Trott and A. J. Olson, Journal of Computational Chemistry, 31 (2010), 455–46,doi: 10.1002/jcc.21334.
CrossRef - Stephanus Daniel Handoko.;Xuchang Ouyang.;Chinh Tran To Su.; CheeKeongKwoh.;Yew Soon Ong. Trans Comput Biol Bioinform,2012 ,(5),126672,doi:10.1109/TCBB.2012.82
CrossRef - Eberhardt, J.; Santos-Martins, D.; Tillack ,A.F.; Forli, S. Journal of chemical information and modeling. 2021, 61, 3891-3898
CrossRef - Sun, YI.; Oberley, L.W.; Li Y. Clinical chemistry. 1988, 34, 497-500
CrossRef - Klaus R. Huber .; Rajagopalan Sridhar.; Elizabeth H. Griffith .; Elmer L. Amma .; Joseph Roberts.Biochimica et Biophysica Acta 915 (1987) 267-276, doi: 10.1016/0167-4838(87)90309-8
CrossRef - Hamid reza ghorbani, hosein attar, ali akbar safekordi, s. Mahdi rezayat sorkhabadi, Asian Journal of Chemistry; Vol. 23, No. 11 (2011), 5111-5118.
CrossRef - Hamid Reza Ghorbani, Iman Fazeli , Ali Asghar Fallahi, Orient. J. Chem., Vol. 31(1), 515-517 (2015), http://dx.doi.org/10.13005/ojc/310163.
CrossRef - Bahareh Khodashenas1 , Hamid Reza Ghorbani , IET Nanobiotechnol., 2016, 10, 3, 158–161, doi: 10.1049/iet-nbt.2015.0062.
- Hamid Reza Ghorbani ,Mazaher Molaei, Mater. Res. Express ,4 (2017) 065017, https://doi.org/10.1088/2053-1591/aa75ba.
CrossRef - Hamid Reza Ghorbani, IET Nanobiotechnol., 2014,8, 4,263–266
doi: 10.1049/iet-nbt.2013.0039.
CrossRef - Maria Zahida .;Nazia Yasmina .; Muhammad Naeem Ashiqb .; Muhammad Safdarc.; Misbah Mirzaa.; Physica B: Physics of Condensed Matter, 2021,624, 413359, doi:10.1016/j.physb.2021.413359.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.










