Design, Synthesis and Anticancer Properties of Novel Hydrazino-Fused Pyrimidines
Sathish Kumar Mittapalli1* , Iffath Rizwana2
, Iffath Rizwana2 , Ch. Hari Prasad Murthy3
, Ch. Hari Prasad Murthy3 , Nimisha Jain1
, Nimisha Jain1 , Sagar Pamu1
, Sagar Pamu1 and Pawan Kumar Gupta1
and Pawan Kumar Gupta1
1Department of Pharmaceutical Chemistry, Amity Institute of Pharmacy, Amity University, Gwalior, Madhya Pradesh, India.
2Department of Pharmaceutical Chemistry, Deccan School of Pharmacy, Hyderabad, Telangana-500081, India.
3Department of Pharmacology, Sri Venkateshwera College of Pharmacy, Hyderabad, Telangana-500081, India.
Corresponding Author E-mail: sattisuma@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/390503
Article Received on : 04 Sep 2023
Article Accepted on : 07 Oct 2023
Article Published : 20 Oct 2023
Reviewed by: Dr.V.Anitha Kumari
Second Review by: Dr. Bapu Thorat
Final Approval by: Dr. Tanay Pramanik
The Pyrimidine system has received great attention and a vital component of genetic material emerged has fundamental source to fight against cancer. The pyrazolo(1,5-a) pyrimidines (5a-5j) were designed based on the structural features of antitumor antimetabolites, synthesized and chemical structures were confirmed using spectroscopic methods such as IR, 1H NMR, 13C NMR, Mass Spectral and elemental analysis. The cytotoxic activity was evaluated by DPPH free radical scavenging assay against standard ascorbic acid and MTT assay against MCF-7, HepG-2, and imatinib as standard. The DPPH assay indicated 5b, 5c, 5e, 5h and 5j were efficient antioxidants, while the MTT assay discloses potent cytotoxicity of 5b, 5d against MCF-7 with 16.61, 19.67µg/ml and 5c, 5h against HepG-2 with 14.32 and 19.24µg/ml compared to 5-FU. The ligands 5c and 5h demonstrated promising towards tyrosine kinase and cyclin dependent kinase 2, respectively and the bonding energy is similar as doxorubicin. Concluding that the compounds had reasonable cytotoxic potential and good association observed between in vitro and in silico studies.
KEYWORDS:Cdks; Drug-Likeness; Molecular Docking Analysis; Pyrimidine; Tyrosine Kinase
Download this article as:| Copy the following to cite this article: Mittapalli S. K, Rizwana I, Murthy C. H. P, Jain. Design, Synthesis and Anticancer Properties of Novel Hydrazino-Fused Pyrimidines. Orient J Chem 2023;39(5). |
| Copy the following to cite this URL: Mittapalli S. K, Rizwana I, Murthy C. H. P, Jain. Design, Synthesis and Anticancer Properties of Novel Hydrazino-Fused Pyrimidines. Orient J Chem 2023;39(5). Available from: https://bit.ly/3tGRIop |
Introduction
Protein kinases have emerged as a possible therapeutic targets, with nearly 30 distinct kinase targets being developed for the phase-I trials1. The cell cycle regulation and proliferation mainly depend on cyclin-dependent kinases (CDKs). The many biochemical targets are deregulated and there are three approved CDKIs like palbociclib, ribociclib and abemaciclib2. Because they are crucial in regulating growth factor signaling, tyrosine kinases are a particularly crucial target and many oncogenic tyrosine kinases catalytic domains ATP binding site is the prime site for the inhibitors3. Gefitinib4, lapatinib5 is a low molecular weight EGFR/erbB2-tyrosine kinase dual inhibitor that competes with the ATP for blockade of the catalytic domain. Purine is a heterocyclic nucleus present in various antimetabolites and modified at positions 2, 4 and 9 in the creation of protein kinase inhibitors. A quinazoline scaffold with a phenylamino pyrimidine moiety is one of the structural characteristics6. Pyrazolo-pyrimidine is a bio isostere of purine7 was given much consideration while creating anticancer scaffolds, as evidenced by PKI-1668 and erlotinib9.
Over the past few decades, multifunctional compounds have been employed in treating multi-factorial disorders like cancer. These compounds are generically classed as hybrid and chimerical medications comprising two or more merged pharmacophore groups permitting binding to two or more targets10. The rational construction of novel “smart” molecules that keep the pre-selected qualities of the original templates is made possible by combining two or more overlapping pharmacophore groups on the same scaffold. This process also has certain advantages, such as a decreased risk of toxicity and unpleasant responses11, 12.
Methotrexate, a competitive inhibitor of DHFR that converts DHF to THF, was the foundation for developing the novel pyrazolo(3,4-d)pyrimidines13. Recently reported as cytotoxic on BEAS-2B (lung carcinoma) cells, Pemetrexed (PMX) is a dihydropyrrolo[2,3-d]pyrimidin nucleus with ethylene spacer linkage to PABA moiety and acts as a folate antagonist. Studies revealed that the greater affinity complexation of WP6A and ATP favouring competitive replacement of PMX was confirmed by NMR spectroscopy5. A folate antagonist previously reported to be an oseltamivir-comparable prospective drug effective against seasonal influenza viruses suppressed SARS-CoV-2 proliferation in Calu-3 cells 14, 15.
 |
Figure 1: The features of antimetabolites’ structures |
The epidermal growth factor (EGF) which regulates epithelial cells, the origin of all carcinomas, and the corresponding cell surface receptor are over expressed in tumours and overproduced with mutant hyperactive variants of EGFR being specifically implicated in the promotion of metastasis and tumour growth. A low molecular weight EGFR/erbB2-tyrosine kinase dual inhibitor, gefitinib4 and lapatinib5, competes with ATP to block the catalytic domain. Purine is a heterocyclic nucleus found in many antimetabolites and its structural properties include a quinazoline scaffold with a phenylamino pyrimidine moiety and different substituent’s at the various positions in the design of protein kinase inhibitors6. PKI-1668, erlotinib9, imatinib16, BIRB79617 and BAY43-900618 received significant attention in the development of scaffolds as anticancer medicines.
A series of pyrazolopyrimidines were discloses that antitumour potential against MCF-7, A549 and HT-29 and docking scores reported with strong bonding affinities against target EGFR-Tyrosine kinase and VEGFR19. Novel Pyrazolo[3,4-d] pyrimidines made by fusion involving 5-aminopyrazoles and cyclic lactams with carbon-aryl(heteryl)idene moieties. The target molecules were synthesised via bioisosterism, with a pyrazole ring serving as a significant substitute and screened for five human cancer cell lines with fifteen analogues, demonstrated the greatest activity, reported by trimethoxy-benzylidene-tetrahydro-pyrazolopyrimidine20.
Gefitinib and erlotinib are EGFR-tyrosine kinase inhibitors generated by the bioisosteric substitution of carbon with nitrogen next to the quinazoline system’s pyrimidine. Piritrexim is a pyridopyrimine analogue that has been shown to be valuable in bladder cancer via the DHFRI mechanism21. Palbociclib demonstrated progression-free survival in metastatic breast cancer patients via the EGRF-TK pathway, while pipemidic acid shown possible antibacterial effects via interactions with translation metals, shows some of the reference compounds that include pyridopyrimidine and pyrazolopyrimidine scaffolds 22, 23. The SRC receptor-tyrosine kinase is overexpressed and active in many human cancers, and it has been related to cancer growth and progression to distant metastases. These findings have resulted in the recent targeting of c-SRC for the development of anticancer medicines, which show promise as a potential cancer therapy pathway24. CDKs are a kind of protein that regulates the stages of the cell cycle, namely cell division, gene transcription, and other key metabolic activities and inhibiting these protein regulators is an important strategy in cancer treatment. A novel series of arylidene-hydrazinyl-thiazole derivatives were created based on the structure-activity connection by changing the substituent in the hydrophobic head and tail of the synthesised compounds. When compared to the reference compound, roscovitine, all the synthesised compounds were shown to reduce proliferation and cause cytotoxicity in HepG2, MCF-7, and HCT-116 cell lines to varying degrees25.
Molecular docking is a bioinformatics-based theoretical simulation technique that uses a computer platform to examine the interaction of molecules (such as ligands and receptors) and anticipate their binding modes and affinity. This strategy has been approved by scientists as a promising means in medicinal chemistry, such as structure-based rational drug design26. Based on all survey, based on all the molecular and structural parameters of some anticancer antimetabolites, we designed novel substituted-hydrazine-fused-pyrimidine analogues and synthesized for development of cytotoxic potential drug candidates.
 |
Figure 2: Inhibitors of the EGFR/erbB2-tyrosine kinase |
Materials and Methods
Materials
The target proteins were downloaded from the PDB27 server and Chemdraw Ultra was used to draw the ligand structures. SwissADME28 utilized to predict the molecular parameters oand molinspiration was one of the web servers to produce the bioactivity scores29. Molecular docking simulations were performed by the tool Schrodinger 20.330 and the discovery studio visualized by Biovia31. All the chemicals availed from Aldrich & SD Fine Chemicals and M.Ps were found using an open capillary tube (Stuart Scientific SMP1). Ethyl acetate and n-butanol (20:80) were used in TLC. The infrared spectra in KBr (Vmax in cm-1) were captured using a Shimadzu-FT-IR infrared spectrometer. 13C(100-MHz) and 1H(400-MHz) NMR spectra using Bruker DPX-300 spectrometers using TMS as an internal standard. Thermo Finnigan LCQ Ion Trap & Agilent-1200 series LC instruments were used to collect high-resolution mass spectra (HRMS). In order to perform the elemental analysis, VARIO EL-III was employed.
Evaluation of the molecular descriptive characteristics
In order to anticipate the drug-like properties, which is the key step in the development of active molecule, the SwissADME online program was used28. The main characteristics of a drug candidate are molecular weight, absorption, a limited amount of molecular flexibility and TPSA32. It is possible to assess a compound permeability and bioavailability using straight forward molecular information33. Based on molecular properties using lipinski’s rule of five criteria states that a molecule is a drug like property if it has five-hydrogen bond donors, ten- hydrogen bond acceptors, five-partition coefficients logP, and the molecular weight of 500 daltons34.
Molecular docking analysis
The crystal structures of tyrosine kinase and cyclin-dependent kinases were obtained from the PDB (ID: 7QHL & 2SRC)35. The protein structure was generated using the protein synthesis programme from the Schrodinger suite 20.3. The protein preparation wizard allowed for the structure’s energy minimization, the insertion of the missing atoms, the elimination of water situated less than 5 from the binding pocket, and the addition of the missing atoms. The grid was made by identifying the active position of the co-crystal and creating a grid box in its centre. The ligands were drawn and the energy was minimised before being created via the Lig-Prep Schrodinger module to produce different conformers. Glide in standard precision mode was used to dock the conformers and the poses were sorted and the top pose was identified. It was known that previous reports stated that co-crystal of target CDK receptor binding modes that the three essential hydrogen binding interactions at active site besides hydrophobic interactions are GLU81, ASP86, and LEU83, and validation was confirmed by redocking of co-crystallized structure with target kinase.
 |
Scheme 1: Representative Scheme for the Synthesis of N4-Chlorophenyl-Hydrazino-Pyrazolopyrimidines |
General Procedure for synthesis of Pyazolo(3,4-d)Pyrimidines
Weigh about 12.77 g (0.1 mol) of 4-Chloro aniline (1), transferred to a beaker and 9.8 ml of nitrous acid was added in cool condition the mixture and kept aside for 30 minutes. By adding 100 ml of ice-cold water to the solid material that had precipitated, it was separated and dried to produce 4-chloro benzene diazonium chloride (2). The compound 2 was taken about 8.5 g (0.05 mol), transferred into R.B.flask and was treated with malononitrile in acetic anhydride under reflux for 5 hrs to afford (4-chlorophenyl)carbonohydrazonoyl dicyanide (3). The compound 3 reacts with hydrazine hydrate in ethyl acetate solution under reflux for 4 hrs to afford the compound 4-[2-(4-chlorophenyl)hydrazinyl]-4,5-dihydro-1H-pyrazole-3,5-diamine (4). In the next step, chalcones were prepared using different aldehydes and ketones. They obtained chalcones by treating compound 4 independently under magnetic agitation for 6 hrs in ethanol and drying to afford the compounds 5a-5j.
Biological evaluation
DPPH radical scavenging assay
Oxidative stress is defined as an imbalance between free radical production and antioxidant defence mechanisms. It is linked to a variety of pathophysiological illnesses, including cancer and neurological ailments. Exogenous or endogenous, enzymatic or non-enzymatic antioxidants can minimise or halt cellular damage, largely by scavenging free radicals. The synthesized compounds were examined for their capacity to scavenge free radicals using the DPPH assay method. In order to create a series of dilutions with concentrations ranging from 100 to 200 µg/ml in methanol. The methanolic solution of the synthesized compound (2ml) was added to a 0.003% (w/v) methanolic solution of DPPH (1ml). The mixture was vigorously shaken for five minutes and kept aside for 30 minutes and measured absobance and ascorbic acid served as the reference at 517 nm. The equation used to get the inhibition ratio (%I) for all substances tested is %I = (Ac-As)/Ac x 100, where Ac is the absorbance of the control and As is the absorbance of the sample36, 37.
MTT Assay for Cytotoxic activity
The MTT assay is a colorimetric technique that measures how much mitochondrial succinate dehydrogenase lowers the yellow dye. The study assumed that a certain number of cells existed and that dead cells in the medium did not diminish the amount of tetrazolium dye and the dye penetrates cells and travel to the mitochondria, where it is transformed into formazan and color intensity measured at 570nm.
The MTT assay was done in triplicates of six independent concn’s to predict cell viability and evaluate cell viability in the suspension and trypsinization. The trypan blue test was carried out. The cells were seeded in 96 well plates at a density of 5.0 X 103 cells per well in 100ml culture medium and incubated overnight at 37°C. The hemocytometer was used to count the cells. Old media was removed after incubation and replaced with a new medium containing test samples at 5, 10, 25, 50 and 100 µg/ml concentrations. After 48 hrs, new tetrazolium dye was added and incubated at 37oC for 3hrs before the solution’s absorbance was measured in a DMSO medium at 570nm38, 39. Calculating the growth inhibition percentage involved:
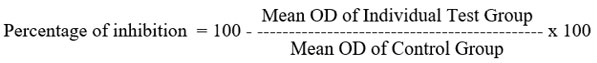
Y = mx+C, was used to get the IC50 value. Here, the viability graph’s Y = 50, M and C values were calculated.
Results and Discussion
Evaluation of the Molecular descriptors
This section lists physical properties40 and except for a few exempt characteristics like Mass, HBD, HBA, Lipophilicity, Molar Refraction, Rotatable bonds, and Lipinski violations. All the compounds were within the allowed ranges; all the information was provided in Table 1.
Table 1: Physicochemical properties of the compounds
|
Title |
Chemical |
M.Wt |
HBD |
HBA |
LogP |
M.R |
TPSA[Ų] |
nrotb |
RO5 |
|
5a |
C22H23ClN6 |
406.91 |
3 |
3 |
3.89 |
133.24 |
78.04 |
6 |
0 |
|
5b |
C22H23ClN6O2 |
438.92 |
5 |
5 |
3.18 |
133.72 |
118.5 |
6 |
0 |
|
5c |
C22H23ClN6O2 |
438.92 |
5 |
5 |
3.18 |
133.72 |
118.5 |
6 |
0 |
|
5d |
C22H22BrClN6 |
485.81 |
3 |
3 |
4.81 |
137.37 |
78.04 |
6 |
1 |
|
5e |
C23H25ClN6O |
436.94 |
3 |
4 |
3.86 |
136.27 |
87.28 |
7 |
1 |
|
5f |
C27H25ClN6 |
468.98 |
3 |
3 |
5.04 |
149.35 |
78.25 |
7 |
1 |
|
5g |
C27H25ClN6O2 |
500.98 |
5 |
5 |
3.98 |
153.24 |
118.52 |
7 |
1 |
|
5h |
C27H25ClN6O2 |
500.98 |
5 |
5 |
3.98 |
153.24 |
118.52 |
7 |
1 |
|
5i |
C27H24BrClN6 |
547.88 |
3 |
3 |
5.6 |
157.05 |
78.25 |
7 |
2 |
|
5j |
C28H27ClN6O |
499.01 |
3 |
4 |
4.7 |
155.85 |
87.27 |
8 |
1 |
“M.Wt = Molecular weight; g/mol; HBD = Hydrogen bond donor; HBA = Hydrogen bond acceptor; lipophilicity (expressed as LogP) LogP = implicit logP method; M.R = Molar Refractivity; TPSA = Topological polar surface area; nrotb = no. of rotatable bonds; RO5 = no. of Lipinski violation”
Bioactivity Score Calculation Using the Molinespiration Toolkit
Calculate the inhibitory activity against various receptor ligands, inhibitors, and enzymes using the molinspiration online toolbox27. Inactivity is expected for scores below -0.50, moderate activity is projected between -0.50 and 0.00 and considerable activity if its bioactivity score is more than 0.0041. All data are shown in Table 2.
Table 2: Molinspiration bioactivity scores prediction.
|
Title |
GPCR ligand |
Ion Channel modulator |
Kinase inhibitor |
Nuclear Receptor ligand |
Protease inhibitor |
Enzyme inhibitor |
|
5a |
-0.09 |
-0.10 |
-0.25 |
-0.52 |
-0.15 |
-0.02 |
|
5b |
-0.14 |
-0.24 |
-0.35 |
-0.68 |
-0.32 |
-0.21 |
|
5c |
-0.24 |
-0.31 |
-0.41 |
-0.69 |
-0.35 |
-0.22 |
|
5d |
-0.27 |
-0.30 |
-0.42 |
-0.87 |
-0.38 |
-0.28 |
|
5e |
-0.3 |
-0.39 |
-0.49 |
-0.85 |
-0.42 |
-0.32 |
|
5f |
-0.1 |
-0.20 |
-0.26 |
-0.54 |
-0.15 |
-0.12 |
|
5g |
-0.11 |
-0.28 |
-0.31 |
-0.64 |
-0.26 |
-0.19 |
|
5h |
-0.19 |
-0.33 |
-0.35 |
-0.65 |
-0.29 |
-0.2 |
|
5i |
-0.22 |
-0.33 |
-0.36 |
-0.79 |
-0.31 |
-0.25 |
|
5j |
-0.25 |
-0.40 |
-0.42 |
-0.78 |
-0.34 |
-0.28 |
Evaluation of the Binding energies to biological targets
All the compounds were targeted for biochemical target was chosen as tyrosine-protein kinase SRC (PDB ID: 2SRC) with single chain A, length 452, and with the co-crystal structure phosphoraminophosphonic acid-adenylate ester with potential docking score -9.758, and Glide model score -139.143. The target as cyclin-dependent kinase-2 (PDB ID: 7QHL), with chain A & C, length 209, and with co-crystal “5-(2-amino-1-ethyl)thio-3-cyclo-butyl-7-[4-(pyrazolo-1-yl)benzyl]amino-pyrazol[4,3-d]pyrimidine” showed potential docking score -10.765 and Glide modelscore -99.351 and all the data was reported in table-3.
The compound 5c reported -7.192 (Figure 3), and 5h reported -7.04K.Cal per mole (Figure 4) against tyrosine kinase, where the standard doxorubicin and pemetrexed reported -8.814K.Cal per mole showed -7.646 K.Cal per mole respectively (Figure 5 & 6). The reference, doxorubicin scored -8.924 K.Cal per mole against CDKs and pemetrexed which reported -8.34 K.Cal per mole (Figures 9 & 10), the compounds 5b and 5h exhibited -6.799 (Figure 7) and -6.915 K.Cal per mole(Figure 8) respectively.
Table 3: Molecular Docking Results targeting 2SRC and 7QHL
|
Compound Code |
2SRC (Tyrosine kinase) |
7QHL (Cyclin Dependent Kinase-2) |
||
|
Glide emodel |
Docking score |
Glide emodel |
Docking score |
|
|
5a |
-54.008 |
-6.099 |
-55.813 |
-6.645 |
|
5b |
-45.255 |
-4.897 |
-61.654 |
-6.799 |
|
5c |
-66.017 |
-7.192 |
-61.081 |
-6.699 |
|
5d |
-59.321 |
-5.973 |
-54.691 |
-6.002 |
|
5e |
-53.516 |
-5.548 |
-52.018 |
-5.915 |
|
5f |
-62.733 |
-6.529 |
-63.604 |
-6.433 |
|
5g |
-59.323 |
-5.641 |
-62.461 |
-5.615 |
|
5h |
-77.117 |
-7.04 |
-71.638 |
-6.915 |
|
5i |
-63.967 |
-5.961 |
-61.406 |
-5.974 |
|
5j |
-62.247 |
-6.3 |
-67.641 |
-6.49 |
|
Doxorubicin |
-95.069 |
-8.814 |
-89.171 |
-8.924 |
|
Pemetrexed |
-84.831 |
-7.646 |
-88.372 |
-8.34 |
|
Co-Crystal |
-139.143 |
-9.758 |
-99.351 |
-10.765 |
 |
Figure 3: 2D and 3D mode affections of 5c against 2SRC |
 |
Figure 4: 2D and 3D mode affections of 5h against 2SRC |
 |
Figure 5: 2D and 3D mode affections of doxorubicin against 2SRC |
 |
Figure 6: 2D and 3D mode affections of Pemetrexed against 2SRC |
 |
Figure 7: 2D and 3D mode affections of 5b against 7QHL |
 |
Figure 8: 2D and 3D mode affections of 5h against 7QHL |
 |
Figure 9: 2D and 3D mode affections of Pemetrexed against 7QHL |
 |
Figure 10: 2D and 3D mode affections of Pemetrexed against 7QHL |
Chemistry
Spectral data of the Synthesized Compounds
[2-(4-chlorophenyl) hydrazinylidene]propanedinitrile (3)
colour: White solid; Yield 85%; Melting point: 165-167˚C; FT-IR (cm-1): 3315 (N-H), 3225(CH–Ar), 1616 (C=N), 1255 (C-N); 1H-NMR: δ 6.69 (m, 2H, J = 8.4, 1.9, 0.6 Hz), 6.91(s,1H, HC=C), 7.38 (m, 2H, J = 8.4, 1.7, 0.6 Hz); 13C NMR: δ 15.3, 18.0, 111.3, 114.1, 145.4, 148.1, 148.4; HRMS: m/z[M+H]+Calculated for C9H5ClN4 204.1711, found 204.1725
4-[2-(4-chlorophenyl)hydrazinyl]-4,5-dihydro-1H-pyrazole-3,5-diamine (4)
Colour: White solid; Yield 88%; Melting point: 220-222˚C; FT-IR (cm-1): 3185 (N-H), 2865 (C-H), 1543 (C=C), 1358 (C-H bending); 1H-NMR: δ 4.15-4.82 (s, 2H, pyrazole), 7.27-7.73 (m, 4H, ArCH, J = 8.0, 7.5, 1.4 Hz), 12.61 (s, 1H, NH); 13C NMR: δ 56.6, 80.5, 117.7, 128.9, 133.7, 148.8, 150.9. HRMS: m/z[M+H]+ Calcd C17H14N4O4 240.35,found 240.25.
3-[2-(4-chlorophenyl)hydrazinyl]-5-methyl-7-[2-phenylethenyl]pyrazolo[1,5-a]pyrimidin-2-amine (5a)
Colour: White solid;Yield 71%; Melting point: 268-270˚C; FT-IR (cm-1): 3035 (N-H), 3055 (Ar-CH), 2755(C-H), 1665(C=N), 1525 (Ar-C=C), 726 (C-Cl); 1H-NMR: δ 1.21 (d, 3H, CH3, J = 6.4 Hz), 1.95 (s, 3H, CH3), 3.51 (d, 1H, J = 9.2, 6.4 Hz), 4.09 (d, 1H, J = 7.9 Hz), 4.62 (d, 1H, J = 7.9 Hz), 5.46 (d, 1H, J = 9.2 Hz), 6.72 (d, 1H, J = 17.3 Hz), 6.88 (d, 1H, J = 17.3 Hz), 7.02-7.25 (m, 3H, J = 8.2 Hz), 7.26-7.52 (m, 6H, ArH, J = 7.6, 1.8, 1.3, 0.5 Hz); 13C-NMR: δ 14.5, 22.2, 46.5, 56.6, 80.5, 117.7, 119.5, 123.8, 126.3, 127.2, 127.8, 128.4, 128.9, 130.3, 133.6, 133.8, 148.8, 151.7; HRMS: m/z[M+H]+ Calculated for C22H23ClN6 406.91, found 406.82; Anal. Calcd C (58.17%), H (6.74%), N (25.47%). Found: C (57.77%), H (6.03%), N (24.84%).
4-[(2-{2-amino-3-[2-(4-chlorophenyl)hydrazinyl]-5-methylpyrazolo[1,5-a]pyrimidin-7-yl}ethenyl]benzene-1,2-diol (5b)
Colour: Brown solid; Yield 68%; Melting point: 275-277˚C; FT-IR (cm-1): 3055 (N-H), 3785 (Ar-CH), 3265 (O-H), 2675 (C-H), 1687 (C=N), 1548 (Ar-C=C), 786 (C-Cl); 1H-NMR: δ 1.21 (d, 3H, CH3, J = 6.4 Hz), 1.95 (s, 3H, CH3), 3.45 (1H, dq, J = 9.0, 6.4 Hz), 4.09 (1H, d, J = 7.9 Hz), 4.62 (d, 1H, J = 7.9 Hz), 5.39 (d, 1H, J = 9.0 Hz), 6.55-6.72 (m, 2H, CH, J = 15.5 Hz), 6.86 (d, 1H, J = 15.5 Hz), 7.02-7.16 (m, 3H, ArH, J = 8.6, 8.1 Hz), 7.28-7.52 (3H, 7.34 (m, J = 8.1, 2.5 Hz); 13C NMR: δ 14.5, 22.2, 46.5, 56.6, 80.5, 115.1, 117.7, 119.5, 121.3, 123.8, 126.9, 128.4, 128.9, 129.6, 133.6, 133.8, 133.7, 133.7, 145.6 , 146.8, 148.8, 151.7; HRMS: m/z[M+H]+ Calculated for C22H23ClN6O2 438.92, found 438.54; Clalcd C (68.70%), H (5.77%), N (18.49%). Found: C (67.54%), H (5.19%), N (18.10%).
4-[2-{2-amino-3-[2-(4-chlorophenyl)hydrazinyl]-5-methylpyrazolo[1,5-a]pyrimidin-7-yl}ethenyl]benzene-1,3-diol (5c)
Colour: Light brown solid; Yield 82%; Melting point: above 300˚C; FT-IR (cm-1): 3045 (N-H, str.), 3146 (Ar-CH), 3355 (O-H), 2645 (C-H), 1669 (C=N), 685 (C-Cl); 1H-NMR: δ 2.71 (s,3H, CH3), 3.78-3.88 (s, 6H, OCH3, J = 8.4, 0.5 Hz), 6.97-7.29 (m, 4H, ArCH, J = 8.4, 1.8 Hz), 7.08-7.96 (m, 3H, ArH, J = 1.8, 0.5 Hz), 8.62 (s, 1H, CH); 13C-NMR: δ 26.7, 56.0, 56.0, 109.1, 111.2, 115.7, 128.0, 128.1, 128.3, 128.2, 130.1, 131.9, 137.7, 148.2, 148.4, 157.3, 159.0; HRMS: m/z[M+H]+ Calculated for C22H23ClN6O2 438.92, found 438.54; Anal. Calcd for: C (65.74%), H (6.78%), N (13.51%). Found: C (65.54%), H (5.80%), N (13.05%).
7-[2-(4-bromophenyl)ethenyl]-3-[2-(4-chlorophenyl)hydrazinyl]-5-methylpyrazolo[1,5-a]pyrimidin-2-amine (5d)
Colour: Light yellow solid; Yield 62%; Melting point: above 300˚C; FT-IR (cm-1): 2954 (N-H), 3167 (Ar-CH), 2875 (C-H), 1578 (C=N), 824 (C-Cl), 525 (C-Br); 1H-NMR: δ 2.71 (s, 3H, CH3), 6.97-7.33 (m, 4H, ArH, J = 8.4, 1.8 Hz), 7.08-7.96 (m, 4H, ArH, J = 1.8, 0.5 Hz), 8.71 (s, 1H, CH), 9.58 (s, 1H, NH), 11.48 (s, 1H, OH), 13.19 (s, 1H, NH); 13C-NMR: δ 26.7, 115.7, 116.8, 118.9, 128.1-128.3, 128.2, 128.2, 128.2, 128.4, 129.4, 131.9, 132.5, 137.7, 157.3, 161.1, 162.2; HRMS: m/z [M+H]+ Calculated for C22H22BrClN6 485.44, found 485.93; Anal. Calcd: C (66.71%), H (5.48%), N (15.47%). Found: C (66.12%), H (4.10%), N (14.81%).
7-[2-(2-methoxyphenyl)ethenyl]-3-[2-(4-chlorophenyl)hydrazinyl]-5-methylpyrazolo[1,5-a]pyrimidin-2-amine (5e)
Colour: Cream solid; Yield 52%; Melting point: above 300˚C; FT-IR (cm-1): 3045 (N-H), 3052 (Ar-CH), 2755 (C-H), 1654 (C=N), 1011 (C-O), 758 (C-Cl); 1H-NMR: δ 2.75 (s, 3H, CH3), 6.97-7.33 (m, 4H, ArH, J = 8.4, 1.8 Hz), 7.08-7.96 (m, 4H, ArH, J = 1.8, 0.5 Hz), 8.71 (s, 1H, CH), 8.58 (s, 1H, NH), 11.47 (s, 1H, OH), 13.18 (s, 1H, NH); 13C-NMR: δ 26.7, 115.7, 116.8, 118.9, 128.1-128.3, 128.2, 128.2, 128.2, 128.4, 129.4, 131.9, 132.5, 137.7, 157.3, 161.1, 162.2; HRMS: m/z [M+H]+ Calculated for C23H25ClN6O 436.44, found 436.93; Anal. Calcd: C (60.48%), H (4.10%), N (18.71%). Found: C (60.48%), H (4.18%), N (18.08%).
3-[2-(4-chlorophenyl)hydrazinyl]-5-phenyl-7-[(Z)-2-phenylethenyl]pyrazolo[1,5-a]pyrimidin-2-amine (5f)
Colour: Brown solid;Yield 75%; Melting point:Melting point: above 300˚C; FT-IR (cm-1): 3155 (N-H), 3175 (Ar-CH), 2784 (C-H), 1555 (C=N), 759 (C-Cl); 1H-NMR: δ 2.73 (s,3H, CH3), 3.78-3.98 (s, 5H, OCH3, J = 8.4, 0.4 Hz), 6.97-7.29 (m, 4H, ArCH, J = 8.4, 1.8 Hz), 7.08-7.97 (m, 3H, ArH, J = 1.8, 0.5 Hz), 8.65 (s, 1H, CH), 9.58 (s, 1H, NH); 13C-NMR: δ 26.7, 56.0, 56.0, 109.1, 111.2, 115.7, 128.0, 128.1, 128.3, 128.2, 130.1, 131.9, 137.7, 148.2, 148.4, 157.3, 159.0; HRMS: m/z [M+H]+ Calculated for C27H25ClN6O2 468.98, found 468.41; Anal. Calcd: C (61.47%), H (6.48%), N (12.41%). Found: C (61.14%), H (5.81%), N (13.15%).
4-[(2-{2-amino-3-[2-(4-chlorophenyl)hydrazinyl]-5- phenylpyrazolo[1,5-a]pyrimidin-7-yl}ethenyl]benzene-1,2-diol (5g)
Colour: Yellow solid; Yield 64%; Melting point: above 300˚C; FT-IR (cm-1): 3154 (N-H), 3106 (Ar-CH), 3284(O-H), 2851(C-H ), 1671(C=N), 1549 (Ar-C=C), 596 (C-Cl); 1H-NMR: δ 2.81 (s, 3H, CH3), 6.91-7.33 (m, 4H, ArH, J = 8.4, 1.8 Hz), 7.08-7.96 (m, 4H, ArH, J = 1.8, 0.5 Hz), 8.71 (s, 1H, CH), 9.58 (s, 1H, NH), 13.18 (s, 1H, NH), 13.19 (s, 1H, NH); 13C-NMR: δ 26.7, 115.7, 116.8, 118.9, 128.1-128.3, 128.2, 128.2, 128.2, 128.4, 129.4, 131.9, 132.5, 137.7, 157.3, 161.1, 162.2; HRMS: m/z [M+H]+ Calculated for C27H25ClN6O2 500.38, found 500.18; Anal. Calcd: C (54.23%), H (4.25%), N (12.65%). Found: C (54.08%), H (4.19%), N (12.08%).
4-[2-{2-amino-3-[2-(4-chlorophenyl)hydrazinyl]-5-phenylpyrazolo[1,5-a]pyrimidin-7-yl}ethenyl]benzene-1,3-diol (5h)
Colour: Light yellow solid; Yield 71%; Melting point: above 300˚C; FT-IR (cm-1): 3148 (N-H), 3087 (Ar-CH), 3255 (OH), 2761(C-H), 1685 (C=N), 856 (C-Cl); 1H-NMR: δ 3.78-3.88 (s, 6H, OCH3), 6.98-7.97 (m, 10H, ArH, J = 8.4, 1.8 Hz), 8.6 (s, 1H, CH), 8.63 (s, 1H, CH), 9.58 (s, 1H, NH); 13C-NMR: δ 56.0, 111.2, 115.7, 119.9, 127.8, 128.0, 128.1, 128.3, 128.2, 130.1, 131.9, 134.2, 137.7, 148.2, 148.4, 153.0, 159.0; HRMS: m/z [M+H]+ Calculated for C27H25ClN6O2 500.88, found 500.98; Anal. Calcd: C (70.38%), H (5.64%), N (11.19%). Found: C (70.10%), H (5.18%), N (11.84%).
7-[2-(4-bromophenyl)ethenyl]-3-[2-(4-chlorophenyl)hydrazinyl]-5-phenylpyrazolo[1,5-a]pyrimidin-2-amine (5i)
Colour: White solid; Yield 59%; Melting point: above 300˚C; FT-IR (cm-1): 2967 (N-H), 3185 (Ar-CH), 2853(C-H), 1665 (C=N), 1529 (Ar-C=C), 794 (C-Cl), 585 (C-Br); 1H-NMR: δ 2.71 (s, 3H, CH3), 6.90-7.39 (m, 4H, ArH, J = 8.4, 1.8 Hz), 7.08-7.96 (m, 4H, ArH, J = 1.8, 0.5 Hz), 8.71 (s, 1H, CH), 9.58 (s, 1H, NH), 11.45 (s, 1H, OH), 13.18 (s, 1H, NH), 13.19 (s, 1H, NH); 13C-NMR: δ 26.7, 115.7, 116.8, 118.9, 128.1, 128.3, 128.2, 128.2, 128.4, 129.4, 131.9, 132.5, 137.7, 157.3, 161.1, 162.2; HRMS: m/z [M+H]+ Calculated for C27H24BrClN6 547.88, found 547.14; Anal. Calcd: C (72.49%), (5.17%), N (12.68%). Found: C (72.19%), H (5.10%), N (12.27%).
7-[2-(2-methoxyphenyl)ethenyl]-3-[2-(4-chlorophenyl)hydrazinyl]-5-phenyl pyrazolo[1,5-a]pyrimidin-2-amine (5j)
Colour: Pinkish brown solid; Yield 62%; Melting point: above 300˚C; FT-IR (cm-1): 3017 (N-H), 3084 (Ar-CH), 2784(C-H), 1664 (C=N), 1548 (Ar-C=C), 1087 (C-O-C), 657 (C-Cl); 1H-NMR: δ 2.71 (s, 3H, CH3), 6.90-7.39 (m, 4H, ArH, J = 8.4, 1.8 Hz), 7.08-7.96 (m, 4H, ArH, J = 1.8, 0.5 Hz), 8.71 (s, 1H, CH), 9.42 (s, 1H, NH), 11.45 (s, 1H, OH), 13.18 (s, 1H, NH), 13.19 (s, 1H, NH); 13C-NMR: δ 26.7, 115.7, 116.8, 118.9, 128.1, 128.3, 128.2, 128.2, 128.4, 129.4, 131.9, 132.5, 137.7, 157.3, 161.1, 162.2; HRMS: m/z [M+H]+Calculated for C28H27ClN6O 499.09, found 499.01; Anal. Calcd: C (72.41%), (5.08%), N (12.52%). Found: C (72.03%), H (5.92%), N (12.08%).
4-Chloro aniline (1), upon diazotization reaction in the presence of nitrous acid at 0-5oC to give compound 2 and followed by treatment with malononitrile in acetic anhydride to afford (4-chlorophenyl)carbonohydrazonoyl dicyanide (3), which upon treated with hydrazine hydrate to afford the compound 4-[2-(4-chlorophenyl)hydrazinyl]-4,5-dihydro-1H-pyrazole-3,5-diamine (4), which was condensed with chalcones to afford the compounds 5a-5j, as shown in the scheme 1. The compounds 5a-5j were obtained in moderate to good yields (52-82%) and recrystallized from ethanol and ethyl acetate. The analogues IR spectra showed the existence of C=N bonds in the 1525–1685 cm–1 area, two NH bonds in the 2900–3150 cm–1 region, OH groups in the 3200–3500 cm–1 region, C–Cl bonds in the 560-850 cm–1 region, and C–Br bonds in the 525–585 cm–1 region. In 1H-NMR spectra, a singlet at δ 1.75 to 2.85 ppm assignable for a methyl group, singlet at δ 3.78-3.88 for methoxy protons, singlet for NH protons δ 7.78-8.55 for hydrazine protons and multiplet for protons in aromatic region at δ 7.08-8.5 in 13C-NMR spectra aromatic carbons revealed at a range of 120-155 ppm, and the pyrazolopyrimidine scaffold carbons absorb at 90-160 ppm region. Moreover, the HRMS and elemental analysis were carried out to confirm the novel compounds.
Biological Evaluation
The scavenging effect percentage of the compound 5h at 100, 150, and 200 µg/ml is 51.4, 60.2, and 68.3. The scavenging property of 5b at 100, 150, and 200 µg/ml is 51.5, 62 and 67.8 respectively. Ascorbic acid demonstrated a 98.3% scavenging efficiency at a 200 µg/ml concentration. The good inhibition of 5c showed 52.8, 62.5, and 69.4% at 100, 150 and 200 µg/ml respectively. All the compounds except 5c, 5b, and 5h exhibited lowest inhibition. The electron-donating hydroxyl groups in the compounds 5c, 5b, and 5h exhibited the highest antioxidant activities compared to other compounds. Table 4 displays the RSC as a percentage.
Table 4: Radical Scavenging Activity in DPPH.
|
Compound |
The concentration of the tested compounds (µg/ml) |
||
|
100 |
150 |
200 |
|
|
5a |
12.8 |
18.3 |
27.6 |
|
5b |
51.5 |
62 |
67.8 |
|
5c |
52.8 |
62.5 |
69.4 |
|
5d |
20.7 |
34.8 |
38.7 |
|
5e |
54.8 |
46.4 |
54.7 |
|
5f |
32.5 |
46.8 |
52.6 |
|
5g |
22.5 |
32.4 |
39.4 |
|
5h |
51.4 |
60.2 |
68.3 |
|
5i |
21.7 |
37.7 |
39.1 |
|
5j |
37.3 |
47.5 |
53.9 |
|
Ascorbic acid |
73.3 |
85.6 |
98.3 |
Using the MTT assay, the cytotoxic effects of each substance were examined against the MCF-7 and HepG-2 cell lines. The cytotoxicity was assessed using various concentrations and 5-FU as the standard drug. The consolidated concentrations of compounds with respect to inhibition percentage as well as cell viability against MCF-7 and Hep G2were given in Table 2.For in vitro anticancer investigations, every chemical underwent screening. The cytotoxic evaluations were done MTT assay revealed that the compounds 5b (Graph-1 & table-7) reported as potential against MCF 7 with the IC50, 16.61 followed by 5d with the IC50 19.67 µg/ml, where the standard reference showed 14.34 µg/m and 5c (Graph-2 & table-8) reported as potential against Hep G2 with the IC50, 14.32 µg/ml. This was followed by 5h with the IC50 19.24 µg/ml, where the standard reported 11.36 µg/ml, and all cell viability studies were reported in Table 5.
Table 5: In-vitro anticancer MTT assay.
|
S.No |
Compound |
IC50 (µg) |
|
|
MCF 7 |
Hep G2 |
||
|
1 |
5a |
59.65 |
45.85 |
|
2 |
5b |
16.61 |
22.36 |
|
3 |
5c |
>100 |
14.32 |
|
4 |
5d |
19.67 |
56.82 |
|
5 |
5e |
>100 |
>100 |
|
6 |
5f |
29.31 |
49.25 |
|
7 |
5g |
>100 |
55.37 |
|
8 |
5h |
35.35 |
19.24 |
|
9 |
5i |
39.08 |
>100 |
|
10 |
5j |
24.36 |
34.65 |
|
11 |
5-FU |
14.34 |
11.36 |
Table 6: Cytotoxicity of 5b against MCF-7 at independent concentrations
|
Concn (µg) |
Abs at 570nm |
% Inhbn |
% Viability |
IC50 (µg) |
|
5 |
0.435 |
32.1 |
67.9 |
16.61 |
|
10 |
0.351 |
54.35 |
45.65 |
|
|
25 |
0.221 |
62.73 |
33.25 |
|
|
50 |
0.235 |
77.73 |
22.27 |
|
|
100 |
0.338 |
71.36 |
18.25 |
|
|
Untreated |
0.628 |
0 |
100 |
|
|
Blank |
0 |
0 |
0 |
Table 7: Cytotoxicity of 5-FU against MCF-7
|
Concn (µg) |
Abs at 570nm |
% Inhbn |
% Viability |
IC50 (µg) |
|
5 |
0.586 |
78.09 |
21.91 |
14.32 |
|
10 |
0.435 |
82.24 |
17.76 |
|
|
25 |
0.362 |
64.24 |
35.76 |
|
|
50 |
0.321 |
41.07 |
58.93 |
|
|
100 |
0.376 |
27.27 |
72.73 |
|
|
Untreated |
0.743 |
0 |
100 |
|
|
Blank |
0 |
0 |
0 |
 |
Graph 1: % Viability of 5b against MCF-7 |
 |
Graph 2: % Viability of 5c against HepG-2. |
Conclusion
As a result, many new pyrazolo(1,5-a) pyrimidines (5a–5j) were synthesized with excellent yield and identified using various spectral methods. The DPPH assay demonstrated that 5b, 5c, 5e, 5h and 5j were efficient antioxidants, while the MTT assay discloses potent cytotoxicity of 5b, 5d against MCF-7 with IC50, 16.61, 19.67µg/ml and 5c, 5h against HepG-2 with IC50, 14.32 and 19.24µg/ml compared to the reference 5-FU, which disclosed at 14.34 and 11.36µg/ml. The ligands 5c and 5h demonstrated potential scores towards tyrosine kinase and 5h and 5b against cyclin dependent kinase 2, respectively and the bonding energy is almost similar as reference doxorubicin. The spectrum of activity for 5c, 5b and 5h maybe due to the electron-donating hydroxyl groups being crucial for potency. Concluding that the compounds had reasonable cytotoxic potential and good association observed between in vitro and in silico studies.
Acknowledgments
The Amity University management in Madhya Pradesh has allowed the writers to conduct this study for which the authors are grateful.
Conflict of Interest
There are no competing interests to declare, according to the authors.
References
- Venkanna A, Subedi L, Teli MK, Lama PD, Nangunuri BG, Lee SY, Kim SY, Kim MH. Positioning of an unprecedented spiro [5.5] undeca ring system into kinase inhibitor space. Scientific Reports. 2020, 10(1), 21265.
CrossRef - Zhang M, Zhang L, Hei R, Li X, Cai H, Wu X, Zheng Q, Cai C. CDK inhibitors in cancer therapy, an overview of recent development. American journal of cancer research. 2021, 11(5), 1913.
- Beretta GL, Cassinelli G, Pennati M, Zuco V, Gatti L. Overcoming ABC transporter-mediated multidrug resistance: The dual role of tyrosine kinase inhibitors as multitargeting agents. European journal of medicinal chemistry. 2017, 142, 271-89.
CrossRef - Dhillon S. Gefitinib: a review of its use in adults with advanced non-small cell lung cancer. Targeted oncology. 2015, 10, 153-70.
CrossRef - Li X, Yang C, Wan H, Zhang G, Feng J, Zhang L, Chen X, Zhong D, Lou L, Tao W, Zhang L. Discovery and development of pyrotinib: A novel irreversible EGFR/HER2 dual tyrosine kinase inhibitor with favorable safety profiles for the treatment of breast cancer. European Journal of Pharmaceutical Sciences. 2017, 110, 51-61.
CrossRef - Park H, Lee H, Seok C. High-resolution protein–protein docking by global optimization: recent advances and future challenges. Current opinion in structural biology. 2015, 35, 24-31.
CrossRef - Kovalova M, Havlícek L, Djukic S, Skerlova J, Perina M, Pospísil T, Rezníckova E. Characterization of new highly selective pyrazolo [4, 3-d] pyrimidine inhibitor of CDK7. Biomedicine & Pharmacotherapy. 2023, 161, 114492.
CrossRef - Matada GS, Dhiwar PS, Abbas N, Singh E, Ghara A, Patil R, Raghavendra NM. Pharmacophore modeling, virtual screening, molecular docking and dynamics studies for the discovery of HER2-tyrosine kinase inhibitors: An in-silico approach. Journal of Molecular Structure. 2022, 1257, 132531.
CrossRef - Barr Kumarakulasinghe N, Zanwijk NV, Soo RA. Molecular targeted therapy in the treatment of advanced stage non‐small cell lung cancer (NSCLC). Respirology. 2015, 20(3), 370-8.
CrossRef - Talevi A. Multi-target pharmacology: possibilities and limitations of the “skeleton key approach” from a medicinal chemist perspective. Frontiers in pharmacology. 2015, 6, 205.
CrossRef - Smith EL, Harrington K, Staehr M, Masakayan R, Jones J, Long TJ, Ng KY, Ghoddusi M, Purdon TJ, Wang X, Do T. GPRC5D is a target for the immunotherapy of multiple myeloma with rationally designed CAR T cells. Science translational medicine. 2019, 11(485), 7746.
CrossRef - Kontoyianni M. Docking and virtual screening in drug discovery. Proteomics for drug discovery: Methods and protocols. 2017, 255-66.
CrossRef - Rafique B, Khalid AM, Akhtar K, Jabbar A. Interaction of anticancer drug methotrexate with DNA analyzed by electrochemical and spectroscopic methods. Biosensors and Bioelectronics. 2013, 44, 21-6.
CrossRef - Chen Y, Zhou X. Research progress of mTOR inhibitors. European journal of medicinal chemistry. 2020, 208, 112820.
CrossRef - Bae JY, Lee GE, Park H, Cho J, Kim J, Lee J, Kim K, Kim JI, Park MS. Antiviral efficacy of Pralatrexate against SARS-CoV-2. Biomolecules & Therapeutics. 2021, 29(3), 268.
CrossRef - Estey E, Levine RL, Lowenberg B. Current challenges in clinical development of “targeted therapies”: the case of acute myeloid leukemia. Blood, The Journal of the American Society of Hematology. 2015, 125(16), 2461-6.
CrossRef - Jin X, Mo Q, Zhang Y, Gao Y, Wu Y, Li J, Hao X, Ma D, Gao Q, Chen P. The p38 MAPK inhibitor BIRB796 enhances the antitumor effects of VX680 in cervical cancer. Cancer biology & therapy. 2016, 17(5):566-76.
CrossRef - Murphy EJ, Booth JC, Davrazou F, Port AM, Jones DN. Interactions of Anopheles gambiae odorant-binding proteins with a human-derived repellent: implications for the mode of action of n, n-diethyl-3-methylbenzamide (DEET). Journal of Biological Chemistry. 2013, 288(6), 4475-85.
CrossRef - Abdelgawad MA, Bakr RB, Alkhoja OA, Mohamed WR. Design, synthesis and antitumor activity of novel pyrazolo [3, 4-d] pyrimidine derivatives as EGFR-TK inhibitors. Bioorganic chemistry. 2016, 66, 88-96.
CrossRef - Ruzi Z, Bozorov K, Nie L, Zhao J, Aisa HA. Novel pyrazolo [3, 4-d] pyrimidines as potential anticancer agents: Synthesis, VEGFR-2 inhibition, and mechanisms of action. Biomedicine & Pharmacotherapy. 2022; 156, 113948.
CrossRef - Chan, D.C.; Fu, H.; Forsch, R.A.; Queener, S.F.; Rosowsky, A. Design, synthesis, and antifolate activity of new analogues of piritrexim and other diaminopyrimidine dihydrofolate reductase inhibitors with ω-carboxyalkoxy or ω-carboxy-1-alkynyl substitution in the side chain. Journal of medicinal chemistry. 2005, 48(13), 4420-31.
CrossRef - Rugo HS, Finn RS, Diéras V, Ettl J, Lipatov O, Joy AA, Harbeck N, Castrellon A, Iyer S, Lu DR, Mori A. Palbociclib plus letrozole as first-line therapy in estrogen receptor-positive/human epidermal growth factor receptor 2-negative advanced breast cancer with extended follow-up. Breast cancer research and treatment. 2019, 174, 719-29.
CrossRef - C. Alves P, Rijo P, Bravo C, MM Antunes A, André V. Bioactivity of isostructural hydrogen bonding frameworks built from pipemidic acid metal complexes. Molecules. 2020, 25(10), 2374.
CrossRef - Yeatman TJ. A renaissance for SRC. Nature Reviews Cancer. 2004, 4(6), 470-80.
CrossRef - El-Naggar AM, El-Hashash MA, Elkaeed EB. Eco-friendly sequential one-pot synthesis, molecular docking, and anticancer evaluation of arylidene-hydrazinyl-thiazole derivatives as CDK2 inhibitors. Bioorganic Chemistry. 2021, 108, 104615.
CrossRef - Tao, X.; Huang, Y.; Wang, C.; Chen, F.; Yang, L.; Ling, L.; Che, Z.; Chen, X. Recent developments in molecular docking technology applied in food science: a review. International Journal of Food Science & Technology. 2020, 55(1), 33-45
CrossRef - Burley SK, Bhikadiya C, Bi C, Bittrich S, Chen L, Crichlow GV, Duarte JM, Dutta S, Fayazi M, Feng Z, Flatt JW. RCSB Protein Data Bank: Celebrating 50 years of the PDB with new tools for understanding and visualizing biological macromolecules in 3D. Protein Science. 2022, 31(1), 187-208.
CrossRef - Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific reports. 2017, 7(1), 42717.
CrossRef - Cheminformatics M. Molinspiration. Molinspiration Cheminformatics. 2010.
- Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nature protocols. 2016, 11(5), 905-19.
CrossRef - BIOVIA DS. Discovery studio modeling environment, release 2017, San Diego: DassaultSystèmes, 2016. Available from:(Accessed 1 September 2016). 2016.
- Sharma T, Jana S. Investigation of molecular properties that influence the permeability and oral bioavailability of major β-boswellic acids. European Journal of Drug Metabolism and Pharmacokinetics. 2020, 45, 243-55.
CrossRef - Castillo-Garit JA, Casanola-Martin GM, Le-Thi-Thu H, Barigye SJ. A simple method to predict blood-brain barrier permeability of drug-like compounds using classification trees. Medicinal Chemistry. 2017, 13(7), 664-9.
CrossRef - Attique SA, Hassan M, Usman M, Atif RM, Mahboob S, Al-Ghanim KA, Bilal M, Nawaz MZ. A molecular docking approach to evaluate the pharmacological properties of natural and synthetic treatment candidates for use against hypertension. International journal of environmental research and public health. 2019, 16(6), 923.
CrossRef - Sun Q, Wang Y, Desgrosellier JS. Combined Bcl-2/Src inhibition synergize to deplete stem-like breast cancer cells. Cancer letters. 2019, 457, 40-6.
CrossRef - Ganapathy M, Bhunia S. Nutraceuticals: The new generation therapeutics. Adv Tech Biol Med. 2016, 4(179), 2379-1764.
- Kehrer JP, Klotz LO. Free radicals and related reactive species as mediators of tissue injury and disease: implications for health. Critical reviews in toxicology. 2015, 45(9), 765-98.
CrossRef - Huang S, Wang DI, Zhang S, Huang X, Wang D, Ijaz M, Shi Y. Tunicamycin potentiates paclitaxel-induced apoptosis through inhibition of PI3K/AKT and MAPK pathways in breast cancer. Cancer Chemotherapy and Pharmacology. 2017, 80, 685-96.
CrossRef - Ma M, Feng Y, Zhang SQ, Duan W, Gao L, Yuan B, Xin M. Design, synthesis and biological evaluation of novel selective PI3Kδ inhibitors containing pyridopyrimidine scaffold. Future Medicinal Chemistry. 2023.
CrossRef - Abdou A, Mostafa HM, Abdel-Mawgoud AM. Seven metal-based bi-dentate NO azocoumarine complexes: Synthesis, physicochemical properties, DFT calculations, drug-likeness, in vitro antimicrobial screening and molecular docking analysis. Inorganica Chimica Acta. 2022, 539, 121043.
CrossRef - Akintemi EO, Govender KK, Singh T. A DFT study of the chemical reactivity properties, spectroscopy and bioactivity scores of bioactive flavonols. Computational and Theoretical Chemistry. 2022, 1210, 113658.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









