Determination of Pesticide Residues in four Major Spices using UPLC-MS/MS and Optimized QuEChERS Sample Preparation Workflow
Ramesh Babu Natarajan1,2 , Joby Thomas Kakkassery2*
, Joby Thomas Kakkassery2* , Anaswara Raveendran1
, Anaswara Raveendran1 , Amrutha Ravi1
, Amrutha Ravi1 and Mohit Mohan1
and Mohit Mohan1
1Quality Evaluation Laboratory, Spices Board, Palarivattom, Kochi, Kerala, India.
2Department of Chemistry, St. Thomas College (autonomous), Thrissur (University of Calicut), Kerala, India.
Corresponding Author E-mail: drjobythomask@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/380325
Article Received on : 06-Apr-2022
Article Accepted on :
Article Published : 30 May 2022
Reviewed by: Dr. Sri Mursiti
Second Review by: Dr. Nityanand Singh Maurya
Final Approval by: Dr. S.A. Iqbal
A high sensitivity method for analysis of pesticide residues in four spices, viz. cardamom, cumin, ginger and chillies, using specifically optimized ‘quick, easy, cheap, effective, rugged and safe’ (QuEChERS) sample preparation workflow and UPLC-MS/MS, was developed for 53 pesticides commonly used in the cultivation of these spices. Limits of quantification of 0.01 mg/Kg for all pesticides was achieved in the four spice matrices studied. Matrix effects were evaluated in each spice matrix and were found to be uniformly suppressive, with maximum matrix suppression observed in chillies and cumin, followed by cardamom and ginger, necessitating the use of matrix-matched calibration for each spice. The analytical method was validated as per European Union (EU) SANTE/12682/2019 guidelines. The method was then applied to 20 real samples of each spice collected from Indian markets, and regulatory compliance was evaluated against the maximum residue limits established by EU and Codex Alimentarius Commission.
KEYWORDS:HPL;, Multi-Residue Methods; Mass Spectrometry; Method Validation
Download this article as:| Copy the following to cite this article: Natarajan R. B, Kakkassery J. T, Raveendran A, Ravi A, Mohan M. Determination of Pesticide Residues in four Major Spices using UPLC-MS/MS and Optimized QuEChERS Sample Preparation Workflow. Orient J Chem 2022;38(3). |
| Copy the following to cite this URL: Natarajan R. B, Kakkassery J. T, Raveendran A, Ravi A, Mohan M. Determination of Pesticide Residues in four Major Spices using UPLC-MS/MS and Optimized QuEChERS Sample Preparation Workflow. Orient J Chem 2022;38(3). Available from: https://bit.ly/3GtzQ2F |
Introduction
The use of spices and condiments to add flavour, colour and aroma to food has always been an indispensable culinary requirement all over the world. This global demand is reflected in the world spice trade, which amounted to 2.88 billion US dollars in 20191, and is steadily increasing. In view of ubiquitous global culinary usage and the extent of world spice trade, food safety issues in spices becomes important. The presence of pesticide residues is now considered as one of the principal food safety issues and is internationally regulated in trade by means of stringent maximum residue limits (MRLs). Thus, the need for development of a sensitive, efficient and reliable pesticide residue analytical method in spices merits an important consideration.
Chromatographic techniques coupled to tandem mass spectrometry, typically GC-MS/MS and LC-MS/MS, have now become the de-facto tools for analysis of pesticide residues with the high sensitivity required to ensure compliance with current MRL regulations. Spices are generally considered as difficult matrices for high sensitivity pesticide residue analysis due to their complex chemical composition, which leads to a high amount of matrix coextractives that can potentially interfere with chromatographic separation and mass spectrometric detection and quantification2–5.
The quick, easy, cheap, effective, rugged and safe (QuEChERS) sample preparation technique6, pioneered in 2003, has since evolved into a versatile methodology for pesticide residue analysis in a variety of matrices. Originally this method was applied to fruits and vegetables and found considerable success in covering many matrices and classes of pesticides. Since then, several modifications have been introduced into the classical QuEChERS method which has extended its range of applicability and made it more efficient7–9.
An important issue to be considered in developing pesticide residue analysis methods in spices is their diverse nature. The Codex Committee on Spices and Culinary Herbs (CCSCH) classifies spices into 6 classes, viz. dried fruits and berries (e.g., chillies, black pepper), dried roots and rhizomes (e.g., turmeric, ginger), dried seeds (e.g., cumin, fennel), dried floral parts (e.g., mace, saffron), dried bark (e.g., cinnamon, cassia) and dried leaves (e.g., basil, oregano)10. The widely varying chemical characteristics of different classes of spices has placed constraints on the applicability of QuEChERS methodology, mainly due to the high amount of matrix coextractives present in spice extracts2. Thus, the study of matrix effects is an important consideration in high sensitivity residue analysis using chromatography and mass spectrometric techniques. Also, as the chemical nature of the matrix is different in each class of spices, specific optimizations are required before a sample preparation method for high sensitivity residue analysis can be applied to the different classes.
In LC-MS/MS, matrix effect arises in the electrospray ionisation source (ESI) and usually manifests in the form of signal suppression11. This poses hindrance to reliable identification and quantification of analytes at the sensitivity levels demanded by present regulatory requirements for pesticide residues. Accordingly, assessing and addressing matrix effects is an integral part of method development in pesticide residue analysis.
In this study, development and validation of an analytical method for 53 commonly used pesticides in four commercially important and extensively used spices belonging to different classes, viz. chillies (dried fruit, with high pigmentation), cardamom (dried fruit, with low pigmentation), cumin (dried seed) and ginger (dried rhizome), using UPLC-MS/MS, is documented. Chromatographic and mass spectrometric parameters for 53 pesticides were optimized for response, peak shape and separation. For all spices, a common acetonitrile extraction step based on buffered QuEChERS procedure was optimized. For cleanup of the extracts, different combination of QuEChERS reagents were applied to each spice and optimized to obtain best accuracy and precision in each case. Matrix effects posed by each of the spice matrices in UPLC-MS/MS analysis of residues were assessed. Method validation was performed as per European Union SANTE/12682/2019 guidelines12. The method was then applied to 20 real samples of each spice collected from local markets, and regulatory compliance of these samples were evaluated against the maximum residue limits established by European Union and Codex Alimentarius Commission.
Materials and Methods
Chemicals and reference standards
LC-MS/MS grade Acetonitrile and methanol were obtained from Biosolv, USA. Analytical grade Ammonium formate, formic acid, anhydrous magnesium sulphate (MgSO4), sodium chloride (NaCl), sodium citrate tribasic dihydrate (C6H5Na3O7.2H2O) and sodium citrate dibasic sesquihydrate (C6H5Na2O7.1.5H2O) were obtained from Merck, India. Graphitized carbon black (GCB), primary secondary amine (PSA) and C-18 end-capped bulk sorbent were purchased from Agilent Technologies, USA. Certified reference materials of 53 pesticides, with purity > 95% for all compounds, were procured from Dr. Ehrenstorfer GmbH (Germany). Individual pesticide standard stock solutions of 1000 mg/L of 30 compounds and the intermediate mixed standard solution at 10 mg/L were prepared in acetonitrile and stored at -20°C until analysis. Working solutions of the mixed standard were prepared daily by appropriate serial dilutions.
Instrumentation
For sample preparation, vortex mixer and centrifuges used were from Remi, India. Low volume concentrator was from PCI, India. Pesticide residue analysis was performed using a Waters Xevo TQS Micro UPLC-MS/MS system, USA. For concentration of the final extracts for reconstitution, a low-volume concentration workstation from PCI Analytics, India was used.
Optimization of instrument conditions
Chromatographic separations in UPLC were performed over a C-18 column (Waters XBridge® BEH 2.5mm, 2.1x100mm). Four combinations of UPLC mobile phases were assessed, viz. acetonitrile – water system with and without buffer, and methanol – water system with and without buffer. The buffer system used was 5 mM ammonium formate / 0.1% formic acid. Gradients were optimized to obtain good separation and peak shapes. The chromatographic conditions and operational parameters of the mass spectrometer are summarized in Table 1. Mass spectrometric analysis was done using electrospray ionization (ESI) and multiple reaction monitoring (MRM) with two transitions per compound. The compound-dependant parameters for the 53 pesticides used in the study are given in Table 2.
Table 1: Optimized chromatography and mass spectrometry method parameters.
|
Parameters |
|
|
UPLC |
|
|
Column |
Waters XBridge® BEH C-18 2.5mm, 2.1x100mm |
|
Mobile Phase |
A: Water with 5mM ammonium formate and 0.1% formic acid B: methanol with 5mM ammonium formate and 0.1% formic acid Flow 0.5 ml/min Gradient: Initial A:B 98:2, 5 min A:B 50:50 curve 6, 7 min A:B 40:60 curve 6, 11 min A:b 25:75 curve 6, 14 min A:b 1:99 curve 6, 17 min A:B 98:2 curve 6. Total runtime 21 min.
|
|
MS/MS |
|
|
Capillary voltage |
0.6 kV |
|
Cone voltage |
40 |
|
Desolvation temp. |
600°C |
|
Source gas |
1100 L/hr |
|
Cone gas |
50 L/hr |
Table 2: Optimized retention times (TR) and MS/MS parameters for 53 pesticides.
|
Pesticide |
TR (min) |
Quantifying transition (m/z) |
Qualifying transition (m/z) |
Collision Energy (V) |
Cone Voltage (V) |
|
Acephate |
12.62 |
183.9/142.95 |
183.9/49 |
20/18 |
10 |
|
Acetamiprid |
5.09 |
223/126 |
223/56.1 |
15/20 |
30 |
|
Amectoctardin |
8.53 |
276.16/244.07 |
276.16/168.06 |
24/14 |
16 |
|
Azoxystrobin |
8.6 |
404/329 |
404/372 |
30/25 |
25 |
|
Bifenazate |
9.55 |
301.1/198 |
301.1/170 |
20/10 |
25 |
|
Boscalid |
8.92 |
342.9/139.9 |
342.9/307 |
20/45 |
25 |
|
Buprofezin |
12.45 |
306.1/201 |
306.1/57.4 |
25/10 |
10 |
|
Carbaryl |
6.88 |
202.1/145.1 |
202.1/127.1 |
25/10 |
25 |
|
Carbofuran |
6.48 |
222.11/165.1 |
222.11/123 |
20/10 |
5 |
|
Chlorpyrifos |
13.72 |
349.9/97 |
349.9/198 |
16/16 |
20 |
|
Cyantraniliprole |
7.13 |
475.2/286 |
475.2/444 |
16/16 |
20 |
|
Cycloxydim |
11.95 |
326/180 |
326/280 |
22/16 |
34 |
|
Cyprodinil |
9.58 |
226/93 |
226/108 |
35/25 |
5 |
|
Diazinon |
10.8 |
305.1/169 |
305.1/96.9 |
35/22 |
20 |
|
Dimethenamid |
8.54 |
276/244 |
276/168 |
26/14 |
17 |
|
Emamectin benzoate |
14.48 |
886.6/158 |
886.6/126 |
30/35 |
20 |
|
Ethion |
13.59 |
385/199 |
385/142.9 |
25/10 |
30 |
|
Fenarimol |
9.84 |
331/81 |
331/268 |
30/25 |
20 |
|
Fenbuconazole |
10.35 |
337/70.1 |
337/125 |
30/20 |
15 |
|
Fenhexamid |
9.68 |
301.96/55.18 |
301.96/97.11 |
35/25 |
35 |
|
Fenpyroximat |
14.78 |
422.2/366.1 |
422.2/138.1 |
30/20 |
5 |
|
Flupicolide |
8.98 |
383/172.999 |
383/109.06 |
66/20 |
40 |
|
Flutriafol |
7.57 |
302.1/70.2 |
302.1/123.1 |
20/25 |
15 |
|
Fluxapyroxad |
9.2 |
382.2/362 |
382.2/342 |
20/10 |
20 |
|
Hexaconazole |
11.33 |
314/70.1 |
314/159 |
20/25 |
15 |
|
Imidacloprid |
4.69 |
256.1/209.1 |
256.1/175.1 |
20/15 |
25 |
|
Iprobenfos |
10.37 |
289/91 |
289/205 |
20/10 |
9 |
|
Malathion |
9.08 |
331/127 |
331/99 |
20/15 |
10 |
|
Mandipropamid |
9.04 |
411.8/328.1 |
411.8/125 |
35/15 |
35 |
|
Mehtiocarb |
8.71 |
226/169 |
226/121 |
20/10 |
25 |
|
Metalaxyl |
7.61 |
280.1/220.1 |
280.1/192.1 |
20/15 |
10 |
|
Methamidophos |
0.6 |
142/93.9 |
142/124.9 |
13/13 |
15 |
|
Methoxyfenozide |
9.2 |
369.2/149.1 |
369.2/313.23 |
15/10 |
15/5 |
|
Penthiopyrad |
10.93 |
360.1/177.1 |
360.1/276 |
47/21 |
30 |
|
Phenthoate |
10.52 |
321/79.1 |
321/135 |
40/20 |
9 |
|
Phosalone |
11.42 |
367.9/181.9 |
367.9/110.9 |
42/14 |
12 |
|
Pirimiphos methyl |
10.92 |
306.1/108.1 |
306.1/164.1 |
32/22 |
25 |
|
Procloraz |
11.02 |
375.84/307.92 |
375.84/70.12 |
24/16 |
10 |
|
Profenofos |
12.54 |
372.9/302.6 |
372.9/127.9 |
40/20 |
25 |
|
Pyraclostrobin |
11.33 |
388.1/193.9 |
388.1/163 |
25/12 |
5 |
|
Quinalphos |
10.37 |
299/96.9 |
299/162.9 |
30/24 |
15 |
|
Quinoxyfen |
13.57 |
308/197 |
308/161.9 |
35/30 |
15 |
|
Spinosad A |
11.68 |
732.6/142 |
732.6/98.1 |
35/30 |
35 |
|
Spinosad D |
12.44 |
746.52/142 |
746.52/98.1 |
35/31 |
40 |
|
Spirodiclofen |
14.76 |
411.14/71.16 |
411.14/313.1 |
15/10 |
35 |
|
Spirotetramat |
9.65 |
374/330 |
374/302 |
30/15 |
20 |
|
Tebuconazole |
10.85 |
308/70.1 |
308/125 |
20/35 |
10 |
|
Thiacloprid |
5.54 |
253/126 |
253/90.1 |
35/20 |
40 |
|
Thiodicarb |
7.17 |
355.08/88.1 |
355.08/108.1 |
16 |
17 |
|
Thiophanate |
7.88 |
371/151 |
371/93.1 |
50/22 |
28 |
|
Triadimefon |
9.17 |
294.1/69.3 |
294.1/197.2 |
20/15 |
25 |
|
Triazophos |
9.53 |
314.1/161.9 |
314.1/118.9 |
35/18 |
22 |
|
Trifloxystrobin |
12.11 |
409/186 |
409/145 |
40/16 |
10 |
Selection of samples
For study of matrix effects and method validation, organically cultivated spice samples were selected after screening to confirm that they were free from the pesticides under study. For evaluation of real samples, 20 market samples each for cardamom, cumin, ginger and chillies were collected from local markets in Kochi, Kerala, India.
Sample preparation and optimization
Homogenization of the four spices were performed so as to simulate their typical culinary usage. Cardamom and ginger samples were crushed thoroughly using a kitchen blender before analysis. Cumin and chillies were ground to fine powder and sieved through ASTM 20 (850 mm) mesh before analysis.
The optimized extraction step was same for all four spices, in which 2 g of each spice was soaked in 8 ml water for 30 minutes, and then 10 ml acetonitrile was added, followed by 4 g MgSO4, 1 g each of sodium chloride and sodium citrate tribasic dihydrate, and 0.5 g of sodium citrate dibasic sesquihydrate. The mixture was then vortexed for 30 seconds and centrifuged at 5000 rpm for 5 minutes.
Owing to the diverse chemical nature of the four spice matrices under study, the cleanup step had to be optimized separately for each spice matrix. From the extract 2ml was taken for cleanup, and the optimized combination of chemicals, viz. MgSO4, PSA, C18 sorbent and GCB were added into each spice as detailed in Table 3. The mixture was then vortexed for 30 seconds and centrifuged at 10,000 rpm for 5 minutes. From the cleaned extract, 2 ml was evaporated to dryness and reconstituted with 1 ml methanol, filtered through 0.2-micron nylon 6,6 membrane filter, and 10 ml was injected into UPLC-MS/MS.
Method performance evaluation
Method performance was assessed as per European Union SANTE/12682/2019 guideline12 by determining linearity, matrix effect, limits of quantification (LOQ), accuracy and precision. Linearity was assessed based on the correlation coefficient (R2) of calibration curves with five calibration levels from 0.005 to 0.075 mg/L. Accuracy was assessed using recovery experiments with spike levels of 0.01 and 0.05 mg/Kg. Intra-day, intra-laboratory precision (repeatability) was calculated as relative standard deviation (RSDr) at two spike levels of 0.01 and 0.05 mg/Kg for all four spices, with 5 replicates for each matrix (same instrument, same analyst, same day). Inter-day precision (reproducibility) was calculated as the relative standard deviation (RSDR) at two spike levels of 0.01 and 0.05 mg/Kg for each spice matrix, with each fortification level analysed in triplicate on three non-consecutive days (n = 9). LOQ was taken as the minimum concentration that could be quantified with accuracy and precision in compliance with the validation requirements.
Evaluation of Matrix effects
Matrix-matched calibration curves for 53 pesticides were setup using post-extraction spikes in blank matrix extracts of cardamom, chilli, ginger and cumin at five concentration levels, viz. 0.005, 0.01, 0.025, 0.05, 0.075 mg/L. Solvent-only (methanol) calibration curves at the same concentrations were also prepared for each pesticide. Matrix effect observed for each spice matrix – pesticide combination was then assessed by comparing the slope of the matrix-matched calibration curve with that of the solvent-only calibration curve.
Results and Discussion
Optimization of UPLC-MS/MS Analysis
Of the four combinations of mobile phase studied, methanol-water composition was in general better than acetonitrile-water composition in obtaining good peak shape and resolution. It was also observed that the use of buffers improved the response and peak shapes in general. Thus, methanol-water mobile phase containing ammonium formate / formic acid (5 mM / 0.1%) buffer was finalized as the mobile phase (Table 1). After optimization, the UPLC chromatogram afforded good separation of the analytes under consideration.
In ESI / MRM mode, for each compound analysed, the mass spectrometer selects a parent ion produced from the analyte molecule and then generates daughter ions from this parent ion by collision induced dissociation with nitrogen inside the collision cell, culminating in a highly specific detection process14. Several parameters influence this ionization process, some generic and other specific to each compound being analysed. These parameters were optimized to obtain maximum response for each compound being analysed. The finalized generic mass spectrometric conditions are summarized in Table 1, and the compound specific parameters are shown in Table 2. These parameters were optimized to minimize interference from co-extractives and maximize response for the individual compounds and spice matrices.
Optimization of QuEChERS extraction and cleanup procedure
In the original QuEChERS procedure validated for fruits and vegetables6, the first step involved extraction of samples directly into acetonitrile in presence of 4g MgSO4 and 1g NaCl. However, spices are dry commodities with moisture content in the range of 8-10%, which is much less than the moisture content in fruits and vegetables. So in the present optimization of the extraction process for the four spices, two additional parameters were studied, viz addition of water to the matrix and soaking time. For extraction, the proportion of sample weight (g) to acetonitrile volume (ml) was maintained optimally at 1:5, as it was noted that decreasing the extraction volume below this ratio resulted in inadequate homogenization during the first vortex mixing step, and above this ratio, there was a dilution of analytes in the extract which would detract from the method sensitivity.
It was established initially that without soaking of the matrix, accuracy and precision within acceptable ranges of method validation cannot be obtained for pesticides at trace levels. This is because saturating the dry spice matrices with water improves the penetration of the extraction solvent and facilitates better partitioning of the residues in the matrix to the solvent. Starting with a sample weight of 2 g and 30-minute soaking time, the sample: water ratios (by weight) of 1:2, 1:4, 1:6 and 1:8 were studied. It was seen that at 1:2 ratio, the recoveries of all the compounds were between 32-61%. At 1:4 ratio, the recovery values showed significant increase, to the range of 50-77%, and further increase in the sample: water ratio did not increase the recoveries significantly. These recovery values were obtained without the cleanup step, which was optimized separately.
Increasing the sample weight while keeping the sample: water ratio at 1:4 did not increase recovery significantly but was seen to affect the repeatability. For 2 g sample weight with addition of 8 ml water with soak time of 30 min, overall intra-day repeatability, RSDr (n=5) was between 8.3 – 13.5% for all compounds, but for 5 g sample weight, this was in the range 14 – 19.6%. This is probably because spices contain significant amounts of crude fibre which makes perfect homogenization difficult, and increasing sample weight consequently would decrease the precision. Increasing soak time beyond 30 minutes did not affect recovery or repeatability to any considerable extent. Thus, sample weight of 2 g, with addition of 8 ml water and a soak time of 30 minutes, were found to be optimal for all four spices. The acetonitrile volume used was fixed at 10 ml itself maintaining the sample-solvent ratio at 1:5.
Addition of sodium citrate salts during the extraction step was considered to enhance the recovery of pH sensitive pesticides. Thus, before optimizing the cleanup step, the effect of buffer salts in the extraction efficiency in the four spices was studied. Using the optimized extraction compositions, recovery studies with and without citrate salts showed that for some pesticides, recovery increased considerably in the presence of citrate salts. For diazinon, carbaryl, chlorpyrifos and malathion, recovery values with addition of citrate salts increased by 13, 19, 17 and 24% in cardamom, 17, 18, 14 and 20% in cumin, 18, 25, 13 and 13% in ginger and 15, 12, 10 and 13% in chillies. For fenhexamid, recovery value increased by 19% in chillies. In all other cases, the variation in recovery values was minor, within ±8% for all compounds in all spice matrices. However, it was deemed beneficial to include sodium citrate salts in the extraction step to improve overall method performance.
To optimize the cleanup step, four QuEChERS reagents were considered, viz. MgSO4, PSA, C-18 end capped sorbent and GCB. MgSO4 is used to remove excess water from the extract and thus facilitate recovery of nonpolar residues. PSA contains primary and secondary amino groups that remove acidic interferences from the extracts. GCB acts by reducing pigments from the extracts but are also known to affect recoveries of planar pesticides. C-18 sorbent is used to remove non-polar interferences.
Spices typically have relatively high amounts of non-polar volatile oil content, of varying chemical compositions, in addition to other active chemical compounds. In cardamom the volatile oil content is around 8 – 9%, in ginger 0.7 – 4% and in cumin 2.7 – 4.3%. Chillies have capsaicinoid content, responsible for their pungency, ranging from 2000 – 5000 mg/Kg. The colour in chillies, arising carotenoid content, range from 0.1 – 0.3%, or 1000 – 3000 mg/Kg15, 16. All these factors contribute to matrix co-extractives which can potentially interfere with analytical performance. Also, as soaking spice samples in water was seen to be very important in spices to obtain good recovery and precision, a natural consequence is the increased water content in the extract which has to be addressed to manage the recovery of non-polar pesticides.
In view of these factors, four combinations of cleanup chemicals were studied: (A) 300 mg MgSO4 + 75 mg PSA + 50 mg C18, (B) 300 mg MgSO4 + 75 mg PSA + 50 mg C18 + 20 mg GCB, (C) 300 mg MgSO4 + 75 mg PSA + 75 mg C18 and (D) 300 mg MgSO4 + 75 mg PSA + 75 mg C18 + 20 mg GCB. Each combination (A) to (D) were applied on 5 samples of each of the four spices spiked at 0.01 mg/kg, and recoveries were assessed. Fig. 1 shows the overall recoveries for five representative compounds, viz. imidacloprid, ethion, chlorpyrifos, quinalphos and spirodclofen, obtained for the four cleanup combinations in the four spices studied.
 |
Figure 1: Optimization of cleanup procedures in four spices based on average recovery for spike level 0.01 mg/kg (n=5). |
In all four spices, cleanup increased recoveries of the studied compounds considerably. Using cleanup combination (C), average recoveries were obtained in the range 83.7 – 97.8 for cardamom. Using cleanup combination (D), average recoveries in the range 98.7 – 102.7% were obtained for cumin. Using cleanup combination (B), average recoveries in the range 87.7 – 106.8% were obtained for ginger, and in the range 93.8 – 104.6% were obtained for chillies. As these recovery values were respectively the highest for each spice and were in accordance with acceptable validation criteria, the respective cleanup combinations were taken as optimal for each spice. Thus, the optimized QuEChERS extraction and cleanup workflow, for the spices cardamom, cumin, ginger and chillies, are summarized in Table 3.
Table 3: Optimized extraction and QuEChERS cleanup scheme for cardamom, cumin, ginger and chillies.
|
Process |
Cardamom |
Cumin |
Ginger |
Chillies |
|
Extraction |
|
|
|
|
|
Sample weight (g) |
2 |
2 |
2 |
2 |
|
Add water (ml) / soak time (min) |
8/30 |
8/30 |
8/30 |
8/30 |
|
Add acetonitrile (ml) |
10 |
10 |
10 |
10 |
|
Add MgSO4 anh. (g) |
4 |
4 |
4 |
4 |
|
Add NaCl (g) |
1 |
1 |
1 |
1 |
|
Add Sodium citrate tribasic dihydrate (g) |
1 |
1 |
1 |
1 |
|
Add sodium citrate dibasic sesquihydrate (g) |
1 |
1 |
1 |
1 |
|
|
Vortexed 30 sec, centrifuged 5000 rpm 5 min. |
|||
|
Cleanup |
|
|
|
|
|
Volume taken for cleanup (ml) |
2 |
2 |
2 |
2 |
|
Add PSA (mg) |
75 |
75 |
75 |
75 |
|
Add C18 sorbent (mg) |
75 |
75 |
50 |
50 |
|
Add GCB (mg) |
0 |
20 |
20 |
20 |
|
Add MgSO4 anh (mg) |
300 |
300 |
300 |
300 |
|
|
Vortexed 30 sec, centrifuged 10000 rpm 5 min. |
|||
|
Concentration and reconstitution |
|
|
|
|
|
Cleaned extract evaporated to dryness (ml) |
2 |
2 |
2 |
2 |
|
Reconstituted in 1:1 MeOH:H2O (ml) |
1 |
2 |
2 |
2 |
Matrix effects
The extent of matrix coextractives obtained using the optimized extraction and cleanup steps was studied gravimetrically. When compared to the matrix load in the extract, the optimized cleanup step reduced the matrix load (mg/ml) to a considerable extent: 53.3% in cardamom, 51% in cumin, 50% in ginger and 56.7% in chillies. The results are shown in Fig. 2.
 |
Figure 2: Effect of optimized cleanup on matrix co-extractives as determined by gravimetric analysis. |
In LC-MS/MS, matrix effect (ME) arises in the electrospray ionization (ESI) source and usually manifests in the form of signal suppression13. In calibration curves, signal suppression manifests as lower slopes in matrix matched calibration curves as compared to solvent-only calibration curves. Matrix matched calibration curves were set up using extracts obtained from blank samples of cardamom, cumin, ginger and chillies using the optimized extraction method. Table 4 shows the regression equations and correlation coefficients obtained for 53 compounds studied in each of the four spices.
MEs were calculated using the following equation:
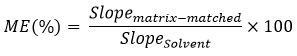
ME between 80-120% are considered negligible, or soft ME, and does not require matrix matched calibration for reliable quantitative results. ME between 50-80% (suppression) and 120-150% (enhancement) are considered medium. ME lower than 50% (suppression) and higher than 150% (enhancement) are considered strong17,18.
The ME posed by the spice matrices were uniformly suppressive and ranged from medium to strong. In cardamom, the ME ranged from 25-80%, in cumin between 10-46%, in ginger between 35-89% and in chillies between 11-67%. Thus, the highest suppression was observed in cumin and chillies. Only 4 pesticides showed matrix suppression in the low ranges (ME > 80%), viz. fenhexamid (88%), fenpyroximat (89%) ad flutirafol (87%) in ginger matrix and pyroaclostrobin (80%) in cardamom matrix. When matrix suppression is low, i.e., ME is between 80 – 100%, results estimated using solvent-only calibration curves will not have large errors. However, with ME < 80%, using solvent-only calibration curves will lead to considerable underestimation of results. As the ME values were > 80% only in 1.8% cases in all the spice – pesticide combinations studied, it was concluded that matrix matched calibration could not be avoided in all four spices so as to obtain reliable results. The matrix effects observed in 53 pesticides analysed in the four spices studied are shown in Fig. 3.
 |
Figure 3: Matrix effects (%) of 53 pesticides investigated in four spices. |
Table 4: Regression equations and correlation coefficient values for pesticides analyzed by LC-MS/MS in solvent (methanol) and in cardamom, cumin, ginger and chillies.
|
Pesticide |
Regression equation, R2 value |
||||
|
Solvent |
Cardamom |
Cumin |
Ginger |
Chillies |
|
|
Acephate |
874x – 233, 0.9952 |
454x – 205, 0.9932 |
192x – 184, 0.9922 |
507x – 182, 0.9902 |
297x – 238, 0.9862 |
|
Acetamiprid |
19728x + 24531, 0.9952 |
13218x + 21588, 0.9912 |
1973x + 19380, 0.9872 |
13218x + 19134, 0.9862 |
6116x + 25022, 0.9912 |
|
Amectoctardin |
22375x – 353, 0.9981 |
9845x – 311, 0.9921 |
5146x – 279, 0.9931 |
14320x – 275, 0.9911 |
9397x – 360, 0.9891 |
|
Azoxystroin |
12353x + 1181, 0.9941 |
7165x + 1040, 0.9881 |
4200x + 933, 0.9881 |
7659x + 922, 0.9921 |
3459x + 1205, 0.9871 |
|
Bifenazate |
23099x – 593, 0.9896 |
15476x – 522, 0.9866 |
7392x – 468, 0.9806 |
12704x – 463, 0.9876 |
15476x – 605, 0.9826 |
|
Boscalid |
3380x – 35, 0.9933 |
2602x – 31, 0.9843 |
1048x – 28, 0.9923 |
1521x – 27, 0.9873 |
777x – 36, 0.9893 |
|
Buprofezin |
49527x – 663, 0.9951 |
33183x – 583, 0.9901 |
13868x – 524, 0.9901 |
17335x – 517, 0.9931 |
17830x – 676, 0.9881 |
|
Carbaryl |
1728x + 34564, 0.9914 |
933x + 30416, 0.9924 |
432x + 27305, 0.9924 |
1158x + 26960, 0.9924 |
639x + 35255, 0.9894 |
|
Carbofuran |
37168x + 1767, 0.9951 |
21558x + 1555, 0.9891 |
13009x + 1396, 0.9941 |
27876x + 1378, 0.9941 |
12266x + 1803, 0.9871 |
|
Chlorpyrifos |
1789x + 10878, 0.9819 |
876x + 9573, 0.9669 |
787x + 8594, 0.9629 |
1180x + 8485, 0.9699 |
751x + 11096, 0.9659 |
|
Cyantraniliprole |
9938x – 569, 0.9988 |
3677x – 501, 0.9918 |
2783x – 450, 0.9908 |
6361x – 444, 0.9898 |
1590x – 580, 0.9958 |
|
Cycloxydim |
8267x – 156, 0.9952 |
3803x – 137, 0.9882 |
1653x – 123, 0.9872 |
4960x – 122, 0.9872 |
1819x – 159, 0.9912 |
|
Cyprodinil |
236x + 11621, 0.9077 |
130x + 10226, 0.9047 |
52x + 9181, 0.9047 |
139x + 9064, 0.8987 |
57x + 11853, 0.9997 |
|
Diazinon |
21039x – 678, 0.9954 |
11151x – 597, 0.9884 |
5049x – 536, 0.9874 |
10309x – 529, 0.9864 |
4839x – 692, 0.9924 |
|
Dimethenamid |
24025x – 365, 0.9979 |
14895x – 321, 0.9909 |
6006x – 288, 0.9909 |
12733x – 285, 0.9939 |
3844x – 372, 0.9939 |
|
Emamectin benzoate |
11650x – 770, 0.9953 |
6291x – 678, 0.9873 |
3961x – 608, 0.9893 |
8388x – 601, 0.9873 |
1980x – 785, 0.9923 |
|
Ethion |
8300x + 19149, 0.9962 |
5312x + 16851, 0.9932 |
3652x + 15127, 0.9912 |
5561x + 14936, 0.9882 |
913x + 19532, 0.9882 |
|
Fenarimol |
856x + 5417, 0.9905 |
368x + 4767, 0.9865 |
300x + 4279, 0.9875 |
531x + 4225, 0.9815 |
385x + 5525, 0.9835 |
|
Fenbuconazole |
17476x + 13519, 0.9911 |
7864x + 11897, 0.9821 |
5592x + 10680, 0.9831 |
11534x + 10545, 0.9831 |
5592x + 13790, 0.9841 |
|
Fenhexamid |
8489x + 7107, 0.9918 |
4584x + 6254, 0.9848 |
2207x + 5615, 0.9858 |
7471x + 5544, 0.9878 |
1358x + 7249, 0.9868 |
|
Fenpyroximat |
10669x + 28873, 0.9993 |
6722x + 25408, 0.9953 |
2667x + 22809, 0.9923 |
9496x + 22521, 0.9963 |
1280x + 29450, 0.9913 |
|
Flupicolide |
14041x + 5096, 0.9953 |
10952x + 4485, 0.9903 |
3370x + 4026, 0.9923 |
10530x + 3975, 0.9913 |
3229x + 5198, 0.9903 |
|
Flutriafol |
30923x + 23741, 0.9974 |
19791x + 20892, 0.9904 |
8040x + 18755, 0.9904 |
26903x + 18518, 0.9934 |
7422x + 24215, 0.9944 |
|
Fluxapyroxad |
18056x + 7566, 0.9939 |
10111x + 6658, 0.9919 |
4153x + 5977, 0.9919 |
13361x + 5901, 0.9919 |
3070x + 7717, 0.9919 |
|
Hexaconazole |
23678x – 789, 0.9934 |
13023x – 694, 0.9924 |
4262x – 623, 0.9884 |
17048x – 615, 0.9904 |
8051x – 805, 0.9884 |
|
Imidacloprid |
15187x – 266, 0.9964 |
10175x – 234, 0.9944 |
3341x – 210, 0.9884 |
9416x – 207, 0.9884 |
5467x – 271, 0.9874 |
|
Iprobenfos |
44698x + 194, 0.9966 |
24584x + 171, 0.9896 |
14303x + 153, 0.9896 |
28607x + 151, 0.9906 |
13856x + 198, 0.9896 |
|
Malathion |
14856x + 1308, 0.9839 |
10102x + 1151, 0.9769 |
6091x + 1033, 0.9819 |
9656x + 1020, 0.9779 |
5348x + 1334, 0.9889 |
|
Mandipropamid |
9253x + 11353, 0.9902 |
5644x + 9990, 0.9862 |
3516x + 8969, 0.9852 |
5089x + 8855, 0.9842 |
1481x + 11580, 0.9872 |
|
Mehtiocarb |
4483x + 33510, 0.9167 |
2331x + 29489, 0.9087 |
1435x + 26473, 0.9147 |
2869x + 26138, 0.9087 |
1524x + 34180, 0.9847 |
|
Metalaxyl |
18905x – 810, 0.9960 |
8318x – 713, 0.993 |
8318x – 640, 0.991 |
10209x – 632, 0.9870 |
6239x – 826, 0.9870 |
|
Methamidophos |
198x + 14601, 0.9639 |
133x + 12849, 0.9629 |
46x + 11535, 0.9569 |
109x + 11389, 0.9609 |
73x + 14893, 0.9869 |
|
Methoxyfenozide |
2731x + 4244, 0.9282 |
1803x + 3734, 0.9232 |
546x + 3353, 0.9202 |
1748x + 3310, 0.9872 |
1038x + 4329, 0.9822 |
|
Penthiopyrad |
8586x + 3839, 0.9966 |
5409x + 3379, 0.9936 |
1631x + 3033, 0.9886 |
5238x + 2995, 0.9956 |
2662x + 3916, 0.9926 |
|
Phenthoate |
9306x + 76635, 0.9586 |
5956x + 67439, 0.9576 |
2419x + 60542, 0.9956 |
4839x + 59776, 0.9896 |
2140x + 78168, 0.9566 |
|
Phosalone |
3230x + 146, 0.9913 |
2003x + 128, 0.9893 |
1421x + 115, 0.9833 |
2519x + 114, 0.9853 |
807x + 149, 0.9843 |
|
Pirimiphos methyl |
23530x – 193, 0.9955 |
13883x – 170, 0.9925 |
2588x – 152, 0.9905 |
17412x – 150, 0.9875 |
6118x – 197, 0.9865 |
|
Procloraz |
7702x + 6803, 0.9918 |
4467x + 5987, 0.9878 |
1848x + 5374, 0.9828 |
5314x + 5306, 0.9898 |
1617x + 6939, 0.9888 |
|
Profenofos |
1226x + 1228, 0.9977 |
883x + 1081, 0.9907 |
282x + 970, 0.9947 |
748x + 958, 0.9887 |
417x + 1253, 0.9917 |
|
Pyraclostrobin |
13534x + 45783, 0.9978 |
10827x + 40289, 0.9938 |
5684x + 36169, 0.9968 |
8662x + 35711, 0.9938 |
2707x + 46699, 0.9908 |
|
Quinalphos |
13845x – 724, 0.9981 |
8861x – 637, 0.9951 |
6369x – 572, 0.9961 |
9138x – 565, 0.9971 |
2354x – 738, 0.9921 |
|
Quinoxyfen |
4550x – 652, 0.9964 |
1137x – 574, 0.9954 |
1592x – 515, 0.9924 |
3367x – 509, 0.9934 |
2002x – 665, 0.9884 |
|
Spinosad A |
34698x – 469, 0.9985 |
12144x – 412, 0.9915 |
7981x – 370, 0.9935 |
22207x – 366, 0.9905 |
14920x – 478, 0.9935 |
|
Spinosad D |
6640x – 6706, 0.9979 |
4249x – 5901, 0.9929 |
1593x – 5298, 0.9899 |
4382x – 5231, 0.9959 |
2722x – 6840, 0.9949 |
|
Spirodiclofen |
3083x + 7175, 0.9915 |
2035x + 6314, 0.9905 |
1356x + 5668, 0.9885 |
2065x + 5597, 0.9855 |
1079x + 7319, 0.9835 |
|
Spirotetramat |
10950x + 7564, 0.9973 |
7008x + 6657, 0.9943 |
4928x + 5976, 0.9963 |
7008x + 5900, 0.9953 |
3723x + 7716, 0.9893 |
|
Tebuconazole |
6278x + 31018, 0.9982 |
2888x + 27296, 0.9902 |
1632x + 24504, 0.9962 |
4144x + 24194, 0.9932 |
3390x + 31638, 0.9902 |
|
Thiacloprid |
45901x – 160, 0.9972 |
29836x – 141, 0.9942 |
12393x – 126, 0.9962 |
36262x – 125, 0.9892 |
18820x – 163, 0.9952 |
|
Thiodicarb |
32888x – 3993, 0.9988 |
14471x – 3514, 0.9918 |
9538x – 3155, 0.9898 |
23351x – 3115, 0.9978 |
7564x – 4073, 0.9928 |
|
Thiophanate |
26577x + 15966, 0.997 |
17807x + 14050, 0.993 |
10897x + 12613, 0.988 |
11960x + 12454, 0.991 |
5050x + 16286, 0.992 |
|
Triadimefon |
11822x + 6412, 0.9933 |
6502x + 5642, 0.9913 |
4138x + 5065, 0.9873 |
7566x + 5001, 0.9873 |
3783x + 6540, 0.9843 |
|
Triazophos |
51525x – 378, 0.9961 |
22671x – 333, 0.9891 |
12366x – 299, 0.9951 |
32461x – 295, 0.9981 |
13396x – 386, 0.9931 |
|
Trifloxystrobin |
17890x + 36481, 0.9988 |
13239x + 32103, 0.9918 |
4830x + 28820, 0.9908 |
12702x + 28455, 0.9938 |
5367x + 37211, 0.9978 |
Method validation
Validation was performed using the optimized sample preparation and instrument parameters as detailed in the respective sections. Good linearity in response was obtained for all 53 compounds with correlation coefficient r2 ≥ 0.99 in solvent and ≥ 0.98 in all four spice matrices, in the calibration range 0.005 to 0.075 mg/L as shown in Table 4. At spiking levels of 0.01 and 0.05 mg/Kg, average recovery values for all compounds obtained were in the range 79 – 114% for cardamom, 83.3 – 102.7% for cumin, 82 – 107.2% for ginger and 90.3-103.6% for chillies.
Intraday precision RSDr values at the spike levels 0.01 and 0.05 mg/Kg (n=5 per level) were in the ranges 10 – 13.2% in cardamom, 6 – 11.3% in cumin, 9.1 – 16.3% in ginger and 4.1 – 7.8% in chillies. The inter-day precision (RSDR) at the same spike levels (n=9 per level) were in the ranges 13.6-17% in cardamom, 9.1-14% in cumin, 14.3-18.7% in ginger and 6.1-9.3% in chillies. The relatively higher RSD values in cardamom and ginger are likely to be due to the higher crude fibre content in these spices, which would lead to a reduction in homogeneity. All the accuracy and precision values were within the acceptability limits of validation parameters, i.e., 70-120% for accuracy and RSD<20% for precision. An LOQ at 0.01 mg/Kg, which was the lowest level studied which gave acceptable accuracy and precision values, was fixed for 53 compounds in all spice matrices. This LOQ is sufficient to address international regulatory requirements like Codex and European Union maximum residue limits (MRLs)19,20.
Application of the method to of real samples
The 20 market samples of each spice analysed using the optimized method showed the presence of residues of typical analytes, namely acetamiprid (0.01 – 0.04 mg/Kg) and quinalphos (0.01 – 0.03 mg/Kg) in cardamom; imidacloprid (0.01 mg/Kg) and profenofos (0.01–0.04 mg/Kg) in cumin; ethion (0.01 mg/Kg), hexaconazole (0.01 mg/Kg) and profenofos (0.01 – 0.04 mg/Kg) in chillies and metalaxyl (0.01 mg/Kg) in ginger. The incidence of residues was highest in cardamom (51%), followed by chillies (33%), cumin (28%) and ginger (8%). Although all the detected pesticides had concentrations less than the extant maximum residue limits of Codex and EU, the results highlight the need of effective residue management and monitoring plans for agrochemicals in these spices.
Conclusion
An efficient and sensitive QuEChERS-based extraction and cleanup workflow was optimized for pesticide residue analysis of four spices belonging to different classes, viz. cardamom (dried fruit, low colour), cumin (dried seeds), ginger (dried rhizome) and chilli (dried fruit, high colour), for 53 commonly used pesticides in the cultivation of these spices, using UPLC- MS/MS. The method used the same buffered acetonitrile extraction procedure for all spices, and cleanup step was individually optimized for each spice. The method was successfully validated as per EU SANTE/12682/2019 guidelines, and an LOQ of 0.01 mg/Kg could be achieved for all pesticides in all four spices. High values of matrix effects observed in all four spices showed that matrix matched calibration is essential for obtaining reliable results. The optimized method can be effectively used in routine analysis of spices in commercial laboratories for assessing compliance with regulatory requirements.
Acknowledgement
The authors gratefully acknowledge the use of instrumentation facilities at Quality Evaluation Laboratory (Kochi, Kerala), Spices Board, Ministry of Commerce, Government of India.
Conflict of Interest
The authors declare that there is no conflict of interest.
Funding Sources
There are no funding source.
References
- OEC world trade data on spices https://oec.world/en/profile/hs92/spices (accessed 2022 -03 -01).
- Vázquez, P. P.; Ferrer, C.; Bueno, M. M.; Fernández-Alba, A. Pesticide Residues in Spices and Herbs: Sample Preparation Methods and Determination by Chromatographic Techniques. TrAC Trends Anal. Chem. 2019 115 (1), 13-12.
CrossRef - Chawla, S.; Patel, H. K.; Gor, H. N.; Vaghela, K. M.; Solanki, P. P.; Shah, P. G. Evaluation of Matrix Effects in Multiresidue Analysis of Pesticide Residues in Vegetables and Spices by LC-MS/MS. J. AOAC Int. 2017, 100 (3), 616–623.
CrossRef - Shabeer, T. A.; Girame, R.; Utture, S.; Oulkar, D.; Banerjee, K.; Ajay, D.; Arimboor, R.; Menon, K. Optimization of Multi-Residue Method for Targeted Screening and Quantitation of 243 Pesticide Residues in Cardamom (Elettaria Cardamomum) by Gas Chromatography Tandem Mass Spectrometry (GC-MS/MS) Analysis. Chemosphere 2018, 193 (1), 447–453.
CrossRef - Girame, R.; Shabeer, T. A.; Ghosh, B.; Hingmire, S.; Natarajan, R.; Dubey, P. N. Multi-Residue Method Validation and Safety Evaluation of Pesticide Residues in Seed Spices Cumin (Cuminum Cyminum) and Coriander (Coriandrum Sativum) by Gas Chromatography Tandem Mass Spectrometry (GC–MS/MS). Food Chem. 2022, 374 (1), 131782.
CrossRef - Anastassiades, M.; Lehotay, S. J.; Štajnbaher, D.; Schenck, F. J. Fast and Easy Multiresidue Method Employing Acetonitrile Extraction/Partitioning and “Dispersive Solid-Phase Extraction” for the Determination of Pesticide Residues in Produce. J. AOAC Int. 2003, 86 (2), 412–431.
CrossRef - Rejczak, T.; Tuzimski, T. A Review of Recent Developments and Trends in the QuEChERS Sample Preparation Approach. Open Chem. 2015, 13 (1), 980–1010.
CrossRef - Lehotay, S. J.; Son, K. A.; Kwon, H.; Koesukwiwat, U.; Fu, W.; Mastovska, K.; Hoh, E.; Leepipatpiboon, N. Comparison of QuEChERS Sample Preparation Methods for the Analysis of Pesticide Residues in Fruits and Vegetables. J. Chromatogr. A 2010, 1217 (16), 2548–2560.
CrossRef - European Committee for Standardization, Foods of Plant Origin – Determination of Pesticide Residues Using GC-MS and/or LC-MS/MS Following Acetonitrile Extraction / Partitioning and Clean-up by Dispersive SPE – QuEChERS Method. Method No. EN 15662:2008 (E), 2008, 1-81.
- FAO/WHO. Codex Committee on Spices and Culinary Herbs (CCSCH) – related standards https://www.fao.org/fao-who-codexalimentarius/committees/committee/related-standards/en/?committee=CCSCH (accessed 2022 -03 -27).
- Furey, A.; Moriarty, M.; Bane, V.; Kinsella, B.; Lehane, M. Ion Suppression; a Critical Review on Causes, Evaluation, Prevention and Applications. Talanta 2013, 115 (1), 104–122.
CrossRef - European Commission. Guidance Document on Analytical Quality Control and Method Validation Procedures for Pesticide Residue Analysis in Food and Feed. Document No. SANTE/12682/2019, 2020, 1-48.
- Kebarle, P.; Verkerk, U. H. Electrospray: From Ions in Solution to Ions in the Gas Phase, What We Know Now. Mass Spectrom. Rev. 2009, 28 (6), 898–917.
CrossRef - Johnson, A. R.; Carlson, E. E. Collision-Induced Dissociation Mass Spectrometry: A Powerful Tool for Natural Product Structure Elucidation, Anal. Chem. 2015, 87 (21), 10668–10678.
CrossRef - Sahana, K.; Nagarajan, S.; Rao, L. J. M. Cumin (Cuminum Cyminum L.) Seed Volatile Oil: Chemistry and Role in Health and Disease Prevention. Chapter 50, Nuts and seeds in health and disease prevention; Elsevier, 2011 (1), pp 417–427.
CrossRef - Arimboor, R.; Natarajan, R. B.; Menon, K. R.; Chandrasekhar, L. P.; Moorkoth, V. Red Pepper (Capsicum Annuum) Carotenoids as a Source of Natural Food Colors: Analysis and Stability—a Review. J. Food Sci. Technol. 2015, 52 (3), 1258–1271.
CrossRef - Liu, C.; Dou, X.; Zhang, L.; Li, Q.; Duan, Y.; Yang, M.; others. Determination of Triazine Herbicides and Their Metabolites in Multiple Medicinal Parts of Traditional Chinese Medicines Using Streamlined Pretreatment and UFLC-ESI-MS/MS. Chemosphere 2018, 190 (1), 103–113.
CrossRef - Fu, Y.; Lu, Q.; Dou, X.; Luo, J.; Yang, M.; others. Matrix-Matched Monitoring Ion Selection Strategy for Improving the Matrix Effect and Qualitative Accuracy in Pesticide Detection Based on UFLC-ESI-MS/MS: A Case of Chrysanthemum. Microchem. J. 2021, 160 (1), 105681.
CrossRef - Codex Alimentarius Commission. Codex MRL Database http://www.fao.org/fao-who-codexalimentarius/standards/pesticide-mrls/en/ (accessed 2022 -02 -01).
- European Commission. The European Pesticide Database https://ec.europa.eu/food/plant/pesticides/eu-pesticides-db_en (accessed 2022 -02 -01).

This work is licensed under a Creative Commons Attribution 4.0 International License.









