Synthesis, Characterization and Evaluation of Biological Activities of Sn(II) Complexes of Schiff Base Incorporating Sulpha Drugs
Kiran Meena1,2 , Virendra Singh Shekhawat2
, Virendra Singh Shekhawat2 , Sarita Varshney2
, Sarita Varshney2 And A K Varshney2*
And A K Varshney2*
1Department of Chemistry, Mohanlal Sukhadia University, Udaipur-313001, Rajasthan, India.
2Department of Chemistry, University of Rajasthan, Jaipur-302004, India.
Corresponding Author E-mail: kiran02meena@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/370617
Article Received on : 25-Jul-2021
Article Accepted on :
Article Published : 23 Nov 2021
Reviewed by: Dr. M. C. Rao
Second Review by: Dr. Selvaraju K
Final Approval by: Dr. Ioana Stanciu
In this study, we report synthesis, characterization and biological activities of four sulpha drug based Schiff base ligands and their Sn(II) complexes. The Schiff bases and their Sn(II) complexes have been synthesized by traditional methods and characterized by the spectral techniques IR, NMR (1H and 13C), mass and TGA-DTA. Newly synthesized Schiff bases (L1-L4) and their Sn(II) complexes (C-1 to C-4) have been screened for antibacterial activity against bacterial strains S. aureus, S. pyogenus, E. coli, P. aeruginosa and antifungal activity against fungal strains C. albicans, A. niger, A. clavatus using broth micro dilution method. Best antimicrobial activity was shown by C-3 complex against E. coli (MIC, 50.0 µg/mL) and A. niger microbial strains (MIC, 100 µg/mL). Moreover, antimalarial activity against plasmodium falciparum was also studied. Complex C-3 was found to be more active against parasite P. falciparum (IC50, 0.04 µg/mL). Results showed that dichloride tin complexes are more active with respect to their corresponding Schiff base ligands.
KEYWORDS:Antibacterial; Antifungal; Antimalarial activities; Sulpha drug Schiff base; Sn(II) complexes
Download this article as:| Copy the following to cite this article: Meena k, Shekhawat V. S, Varshney S, Varshney A. K. Synthesis, Characterization and Evaluation of Biological Activities of Sn(II) Complexes of Schiff Base Incorporating Sulpha Drugs. Orient J Chem 2021;37(6). |
| Copy the following to cite this URL: Meena k, Shekhawat V. S, Varshney S, Varshney A. K. Synthesis, Characterization and Evaluation of Biological Activities of Sn(II) Complexes of Schiff Base Incorporating Sulpha Drugs. Orient J Chem 2021;37(6). Available from: https://bit.ly/3xgyvax |
Introduction
Hugo Schiff in 1864 first reported preparation and coordination properties of Schiff bases. Schiff bases which have azomethine linkage (–CH=N–) can be synthesized by condensation of active carbonyl compounds with primary amines under acidic or basic conditions1-2. The azomethine linkage is responsible for coordination properties and biological activities such as antimalarial3, anticancer4, antibacterial5, antifungal6, antitubercular7, anti-inflammatory8, antitumor9, insecticidal9, catalytic activity10 and other applications11.
Bacterial and fungal infectious diseases increased mortality in living organism. Mainly following bacterial strains E. coli, P. aeruginosa, S. aureus, S. pyogenus are responsible for many diseases like food borne diseases and food poisoning etc. With the time microbes may have developed resistance to antibiotics that causes many difficulties in treatment of such diseases and it has created global concern to human health. Thus, it is urgent necessity to the exploration and development of new more effective antibacterial and antifungal drugs. Schiff base and their complexes has potential to be good antibacterial and antifungal agents12-13.
Malaria is one of the most destructive infectious diseases and causing serious health problems. P. falciparum, P. ovale, P. vivax and P. malaria14 species of plasmodium are responsible for malaria in humans. Among these species P. falciparum parasite is most dangerous and which is vectored by female anopheles mosquito. For the treatment of malaria, first discovered antimalarial drug was quinine, isolated from the bark of cinchona plant15. Currently, chloroquine is used to treat malaria because it is less toxic, cheaper and effective drug16.
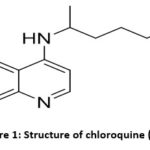 |
Figure 1: Structure of chloroquine (CQ) |
The sulpha drugs containing (–SO2NH–) moiety are extensively used as, antimicrobial17, anti-inflammatory18, antibacterial19, antifungal19, anticancer20, antiviral agents20, diuretics, anticonvulsants, hypoglycaemic, HIV protease inhibitors activity and helpful in both prevention and cure of bacterial infections21-27. Sn(II) compounds have many applications in polymer and dyes industry as catalyst, antitumor and herbicidal use, as photostabilizers, chelating ability, thermal stability, optical nonlinearity28, miticides, molluscides, fungicides, surface disinfectants, wood preservatives29 and marine antifouling30 paints. Methyltetrahydrofuran (MeTHF) is an environmently friendly solvent. It has boiling point 80oC which is higher than boiling point of tetrahydrofurane (THF) 66oC so it increases reaction rate. However, THF is highly miscible with water and generate considerable waste water and waste solvent while MeTHF is less soluble in water, it is low peroxide producing green solvent and it can be easily dried31.
Here in, we describe synthesis, characterization and biological evaluation of various types of sulpha drugs based Schiff base ligands and their Sn(II) complexes using MeTHF as green solvent.
Experimental
Materials and methods
Anisaldehyde, sulphacetamide, sulphamethazine and sulphamerazine were bought from Sigma-Aldrich, India. Sulphadiazine and Anhydrous metal salt (tin chloride) were purchased from Alfa-Aesar. Standard method32 was used for drying the solvents. All the reactions were carried out under anhydrous conditions. Purification of all the compounds has been checked by aluminium silica plate. Melting points of were measured by the Myra melting point apparatus. IR spectrum was recorded using (Bruker alpha-T FT-IR spectrometer) from Department of Chemistry, MLSU, Udaipur (Raj.), India. Spectrum of 1H & 13C-NMR were recorded (BRUKER AVANCE NEO 500 MHz NMR SPECTROMETER) using DMSO-d6 solution from SAIF, Panjab University Chandigarh, India. Mass spectrum was obtained (SYNAPT-XSDBATOF-MS ES+ mass spectrometer) from SAIF, Chandigarh, India. Thermogravimetric analysis was recorded (DTA-TGA Perkin Elmer 6000) from MNIT Jaipur, India. Antimicrobial activities of the synthesized compounds have been performed at Micro Care Laboratory, Surat (Gujarat), India.
Synthesis of ligands
Sulpha drug based Schiff base ligands have been synthesized as per reported method33-34. In this method, we have used a 100 mL neat and clean round bottom flask, 5 mL of alcoholic solution of anisaldehyde (1.22 mL, 0.01mol) was mixed with hot absolute ethanol-acetone solution of sulphacetamide (1.24 g, 0.01 mol) / sulphadiazine (2.50 g, 0.01mol) / sulphamethazine (2.7 g, 0.01mol) / sulphamerazine (2.64 g, 0.01mol) in 1:1 molar ratio. To this solution we have added 4-5 drops of catalyst (glacial acetic acid) and refluxed for about 24 h on oil bath at 60-70oC temperature. Completion of the reaction has been checked by TLC and then it was allowed to cool in refrigerator. The obtained precipitates were filtered off, washed several times with the cold ethanol and purification was done by recrystallization from absolute ethanol. The scheme of the reaction is shown in figure 2.
 |
Figure 2: Synthesis of sulpha drug based Schiff base ligands [L1-L4] |
N-((4-((4-methoxybenzylidene) amino) phenyl)sulfonyl)acetamide [L1]
C16H16N2O4S; Yellow colored; yield, 68.42%; m.p. 210oC; FT-IR (KBr, ѵ cm-1): ѵ ˃C=N (1635 cm-1), ѵ NH (3260 cm-1), ѵ OCH3 (2920 cm-1), ѵ COCH3 (1700-1720 cm-1), ѵ SO21315 & 1150 cm-1(Asy.& Sym.), ѵ C=Caro (1495 cm-1), ѵ =C-H (3050 cm-1). 1H-NMR: δ (ppm)= 8.55 (S, 1H, CH=N), 11.67 (S, 1H, NH), 1.87 (S, 3H, CH3), 3.85 (S, 3H, OCH3), 7.10 (d, 2H, J= 1.8 Hz), 7.87 (dd, 2H, J= 2.0 Hz), 7.38 (d, 2H, J=1.75 Hz), 6.61 (d, 2H, J= 1.75 Hz). 13C-NMR: δ (ppm) =162.38 (–CH=N–), 23.07 (CH3), 55.49 (OCH3), 191.21 (C=O), 135.47 (C1), 121.19 (C2,C6), 123.73(C3, C5), 142.00 (C4), 128.32 (C7), 130.92 (C8, C12), 114.42 (C9, C11), 164.12 (C10) (Ar-C). Mass (m/z): Base peak =332.62 (M)+[C16H16N2O4S]+, 333.67 (M+1) [C16H17N2O4S]+, 107.95 [SO2COCH3]+,cal.332.08; found 332.62.
4-((4-methoxybenzylidene) amino)-N-(pyrimidin-2-yl) benzenesulfonamide [L2]:
C18H16N4O3S; Dark red colored; yield, 50%; m.p. 210–240oC; FT-IR (KBr, ѵ cm-1): ѵ CH=N (1620 cm-1), ѵ NH (3270 cm-1), ѵ SO2 (1320 & 1200 cm-1), ѵ C=Caro (1572 cm-1), ѵ =C-H (2970 cm-1). 1H-NMR: δ (ppm) = 8.52 (S, 1H, CH=N), 11.66 (S, 1H, NH), 3.65 (S, 3H, – OCH3), 6.73 (d, 2H, J= 9.05 Hz), 6.67 (dd, 2H, J= 10 Hz), 8.47 (d, 2H, J= 4.85 Hz), 7.66 (d, 2H, J= 4.85 Hz), 8.52 (d, 2H, J= 4.95 Hz, Hdiazine), 8.27 (t, 1H, J= 5 Hz, Hdiazine). 13C-NMR: δ (ppm) = 158.14 (CH=N), 55.93 (OCH3), 115.59 (C1), 122.40 (C2, C4), 161.06 (C3), 142.99 (C5), 131.46 (C6, C10), 130.19 (C7, C9), 150.99 (C8), 128.40 (C11), 130.20 (C12, C16), 114.40 (C13, C15), 162.90 (C14) (Ar-C). Mass (m/z): 368.60 (M)+[C18H16N4O3S]+, 370.56 (M+2), 372.56 (M+4), 374.58 (M+6), 376.60 (M+8), 378.57 (M+10), 158.88 [C4H4N3SO2]+, Cal. 368.09; found 368.41.
N-(4,6-dimethylpyrimidin-2-yl)-4-((4-methoxybenzylidene) amino) benzenesulfonamide [L3]
C20H20N4O3S; Orange colored; yield, 79%; m.p. 190oC; FT-IR (KBr, ѵ cm-1): ѵ CH=N (1626 cm-1), ѵ NH (3350 cm-1), ѵ CH3 (2855 cm-1), ѵ OCH3 (2950 cm-1), ѵ SO2 (1350 &1247 cm-1). 1H-NMR: δ (ppm) = 8.52 (S, 1H, CH=N), 11.50 (S, 1H, NH), 3.81 (S, 3H, OCH3), 7.07 (d, 2H, J= 2.7 Hz), 7.02 (dd, 2H, J= 5 Hz), 6.78 (d, 2H, J= 8.5 Hz), 7.87 (d, 2H, J= 8.5 Hz), 6.56 (m, 6H, Hdiazine), 2.25 (d, 1H, J= 5 Hz, CH3). 13C-NMR: δ (ppm) = 160.22 (CH=N), 55.30 (OCH3), 22.90 (CH3), 111.65 (C1), 167.17 (C2, C4), 171.90 (C3), 134.03 (C5), 129.80 (C6, C10), 129.30 (C7, C9), 150.90 (C8), 131.70 (C11), 130.19 (C12, C16), 114.40 (C13, C15), 158.02 (C14) (Ar-C). Mass (m/z): 396.74 (M)+[C20H20N4O3S]+, Base peak = 397.77 (M+1) [C20H21N4O3S]+, 398.78 (M+2), 400.81 (M+4), 363.68 [C19H16N4O2S]+, Cal. 396.13; found 396.74.
4-((4-methoxybenzylidene)amino)-N-(4-methylpyrimidin-2-yl)benzenesulfonamide[L4]:
C19H18N4O3S; Brown colored; yield, 64.92%; m.p. 204–215oC; FT-IR (KBr, ѵ cm-1): ѵ CH=N (1590 cm-1), ѵ NH (3320 cm-1), ѵ CH3 (2850 cm-1), ѵ OCH3 (2945 cm-1), ѵ SO2 (1347 & 1243 cm-1). 1H-NMR: δ (ppm)= 8.53 (S, 1H, CH=N), 11.31 (S, 1H, NH), 3.86 (S, 3H, OCH3), 7.08 (d, 2H, J= 8.7 Hz), 8.31 (dd, 2H, J= 1.5 Hz), 7.63 (d, 2H, J= 8.75 Hz), 8.00 (d, 2H, J= 8.5 Hz), 7.86 (d, 1H, J= 1.95 Hz), 6.95 (m, 1H), 2.32 (d, 3H, J= 10 Hz, CH3). 13C-NMR: δ (ppm) = 160.00 (CH=N), 55.57 (OCH3), 23.22 (CH3), 111.91 (C1), 152.84 (C2), 168.90 (C3), 191.18 (C4), 131.68 (C5), 129.36 (C6, C10), 124.82 (C7, C9), 156.80 (C8), 129.91 (C11), 130.78 (C12, C16), 114.44 (C13, C15), 164.10 (C14) (Ar-C). Mass (m/z): 382.53 (M)+[C19H18N4O3S]+, 383.82 (M+1), 384.82 (M+2), Base peak = 248.64[C11H10N3SO2]+, 367.89[C18H15N4O3S] +,cal. 382.11; found 382.44.
Synthesis of Sn(II) Complexes
Tin(II) complexes under investigation were prepared through the reaction of anhydrous tin(II) chloride (0.001mol, 0.18g) with sulpha drug based Schiff base ligands (L1, 0.66 g, 0.002 mol; L2, 0.73 g, 0.002 mol; L3, 0.79 g, 0.002 mol; L4, 0.76 g, 0.002 mol) in MeTHF as reaction medium in 1:2 molar ratio in a 100 mL neat and clean round bottom flask and refluxed on oil bath at 50-65oC for about 4-5 hours. In reaction mixture to adjust pH 5-6 we have added 2-3 drops of glacial acetic acid. The obtained precipitates were filtered off and washed with cold absolute ethanol 2-3 times and kept in refrigerator for 2-3 days for drying.
Purification of the obtained solid compounds was done by recrystallization using absolute ethanol. Purity of each complex was checked by TLC by using silica gel-G as an adsorbent. Structures of synthesized complexes have been shown in figure 3.
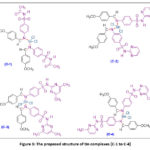 |
Figure 3: The proposed structure of tin complexes (C-1 to C-4) Click here to View figure |
[C33H34Cl2N4O8S2Sn](C-1)
Dark brown colour; yield, 73.77%; m.p. 223oC; FT-IR (KBr, ѵ cm-1): ѵ CH=N (1612-1623 cm-1), ѵ C-O-C (1090 cm-1), ѵ SO2 (1315-1325 cm-1) (Asy.) & 1145-1150 cm-1 (Sym.). 1H-NMR: δ(ppm) = 8.49 (S, 2H, CH=N), 1.05 (S, 6H, CH3), 3.71 (S, 6H, OCH3), 6.87 (d, 4H, J= 3.5 Hz), 7.48 (dd, 4H, J= 5 Hz), 7.66 (d, 4H, J= 8.5 Hz), 6.58 (d, 4H, J= 8.5 Hz). 13C-NMR: δ(ppm) = 169.3 (CH=N), 206.42 (C=O), 55.92 (COCH3), 18.43 (CH3), 137.7 (C1), 116.3 (C2, C6), 127.30 (C3, C5), 130.09 (C4), 122.12 (C7), 123.86 (C8, C12), 114.35 (C9, C11), 168.7 (C10) (Ar-C). Mass (m/z): 868.42(M)+, 870.34 (M+2), 872.80 (M+4), 851.85 [C33H33Cl2N4O7S2Sn]+,904.22 [C33H38Cl2N4O10S2Sn]+, 905.75 [C33H39Cl2N4O10S2Sn], cal. 868.04; found 868.42.
[C36H32Cl2N8O6S2Sn](C-2)
Red colour; yield, 52%; m.p. 252oC; FT-IR (KBr, ѵ cm-1): ѵ (CH=N) (1605 cm-1), ѵ NH (1070 cm-1). 1H-NMR: δ (ppm) = 8.00 (S, 2H, CH=N), 11.66 (S, 2H, NH), 3.81 (S, 6H, OCH3), 6.90-8.00 (m, 16H, Aryl), 8.48 (d, 4H, Hdiazine), 7.02 (t, 2H, Hdiazine). 13C-NMR: δ (ppm) =174.2 (CH=N), 55.80 (OCH3), 169.3 (C1), 157.9 (C2, C4), 115.3 (C3), 139.7 (C5), 127.3 (C6, C10), 125.7 (C7, C9), 147.1 (C8), 122.7 (C11), 124.0 (C12, C16), 114.3 (C13, C15), 160.6 (C14) (Ar-C). Mass (m/z): cal. 926.05; found 926.46.
[C42H44Cl2N8O6S2Sn](C-3)
Orange colour; yield, 79.80%; m.p. 309oC; FT-IR (KBr, ѵ cm-1): ѵ CH=N (1598 cm-1), v NH (1066 cm-1). 1H-NMR: δ (ppm)= 8.50 (S, 2H, CH=N), 10.95 (S, 2H, NH), 3.81 (S, 6H, OCH3), 2.49 (d, 12H, J= 1.5 Hz, CH3diazine), 7.66 (m, 2H, Hdiazine), 7.01 (d, 4H, J= 8.7 Hz), 6.56 (d, 4H, J= 8.7 Hz), 6.55-6.93 (m, 8H, Aryl). 13C-NMR: δ(ppm) = 81.26 (CH=N), 23.02 (CH3), 160.6 (C1), 114.3 (C2, C6), 124.0 (C3, C5), 122.7 (C4), 143.10 (C7), 125.7 (C8, C12), 127.3 (C9, C11), 168.5 (C13), 166.6 (C14, C16), 111.88 (C15), 130.19 (C10) (Ar-C).Mass (m/z): 1010.96 (M)+ [C42H44Cl2N8O6S2Sn]+, 1012.25 [M+2] [C42H46Cl2N8O6S2Sn]+, 1014.58 [M+4][C42H48Cl2N8O6S2Sn]+,995.85 [C41H41Cl2N8O6S2Sn]+, 996.83 [C42H44Cl2N7O6S2Sn]+, 1046.73[C42H48Cl2N8O8S2Sn]+, 1048.88 [C42H50Cl2N8O8S2Sn]+, 1050.94 [C42H52Cl2N8O8S2Sn]+, 1031.30 [C41H41Cl2N8O8S2Sn]+, 1016.85 [C41H46Cl2N8O7S2Sn]+, Cal. 1010.14; found1010.96.
[C38H36Cl2N8O6S2Sn](C-4)
Mud colour; yield, 65.50%; m.p. 310oC; FT-IR (KBr, v cm-1): ѵ CH=N (1585 cm-1), v NH (1066 cm-1). 1H-NMR: δ(ppm) = 8.00 (S, 2H, CH=N), 11.68 (S,2H, NH), 3.81 (S, 6H, OCH3), 2.50 (t, 6H, CH3), 6.93-9.20 (m, 16H, aryl), 8.11 (q, 4H, Hdiazine). 13C-NMR: δ(ppm) = 168.9 (CH=N), 55.8 (OCH3), 23.90 (CH3), 170.1 (C1), 110.1 (C2,C4), 168.9 (C3), 147.1 (C5), 124.0 (C6, C10), 127.3 (C7, C9), 139.7 (C8), 125.7 (C11), 127.3 (C12, C16), 114.3 (C13, C15), 160.6 (C14) (Ar-C). Mass (m/z): Cal. 954.08; found 954.52.
Antibacterial and Antifungal Activity
Antibacterial activity of ligands (L1-L4) as well as their Sn(II) complexes (C-1 to C-4) have studied by Broth dilution method35&36 and MIC values were calculated. Following bacterial strains viz. S. aureus, S. pyogenus (gram positive) and E. coli, P. aeruginosa (gram negative) were used. These strains were procured from institute of microbial technology, Chandigarh, India. Dimethyl sulfoxide solvent was used for serial dilution to receive the appropriate concentration of drugs.
In this method, stock solution of concentration 2000 µg/mL was prepared. In primary screening, the stock solution was diluted to give 1000, 500, 250 µg/mL concentration and screened for their activity and active drug then screened in secondary screening with 200, 100, 50, 25, 12.5, 6.2 µg/mL concentration. After serial dilution the control tube with lowest concentration was taken. The control tubes without antibiotic were immediately subcultured by uniform spreading over a quarter of plate of medium with the help of loop for growth of the test organism and control tubes incubated overnight at 37oC temperature. After incubation, turbidity was measured and microbial growth in the control tubes was compared. The control tubes before incubation represented the original inoculums. The effect of concentration of drug was checked after incubation and the control tube with lowest concentration of drug (lack of turbidity), was shown better inhibition in growth of bacteria. Inoculums size was adjusted to 108Cfu/mL for bacterial test strain by comparing the turbidity in the tubes.
The antifungal activities of the synthesized compounds were tested against fungal strains C. albicans, A. niger, A. clavatus and MIC values were determined. SDB (Sabourand dextrose broth) medium was used for the fungal nutrition, which contains 10 g peptone (from casein), 40 g dextrose (glucose), 1L of distilled water, pH adjusted to 5.6 at 25oC37. Essential amount of the compounds was added into this medium in DMSO. Serial dilutions were prepared for primary and secondary screening as discussed in previous section. The control tubes were kept for incubation for 72 hours at 22oC and MIC were recorded. Standard drug Greseofulvin was used.
Antimalarial Activity
In vitro antimalarial activities of all ligands and their Sn(II) complexes were evaluated against P. falciparum strain. In antimalarial activity we have used 96 well microtitre plates. The cultures of P. falciparum strain were maintained in the medium RPMI 1640 supplemented with 1% D-glucose, 25 Mm HEPS, NaHCO3 0.23% and 10% heat inactivated human serum. In this method, prepared a stock solution of (5 µg/mL) of all test samples using DMSO.
Serial dilutions were prepared with culture medium in same solvent. 20 µL of the samples were diluted at five fold to give 0.4 µg/mL to 100 µg/mL concentration which were kept in the culture plates for incubation at 37oC for 36-40 hour into a candle jar. Thin blood smears were prepared after incubation from each well then stained it with JSB stain. To record maturation of ring stage parasites into trophozoites and schizonts, slides were microscopically examined38-43 and percent inhibition in maturation, average rings of trophozoites and schizontes were recorded per 100 parasities from the duplicate wells. Results were expressed as IC50 value as depicted in table 1.
Results and Discussion
Preparation and Characterization of Sn(II) complexes
[Sn(L)2Cl2] type of complexes were prepared through the reaction of anhydrous tin dichloride with bi-dentate sulpha drug ligands in 1:2 molar ratio using three different solvents ethanol, MeTHF and THF were used and MeTHF solvent was found with best results. The reaction found to be quite facile and could be completed in 4-5 h of refluxing on oil bath. All newly synthesized complexes were obtained in the form of colored solid and the color of the complexes are associated with the presence of chromophoric azomethine group (˃C=N–). FT-IR gives the information about functional group attached to metal atom44. In FT-IR spectrum of all the complexes azomethine peak shifted towards lower frequency in Sn(II) complexes that represent bonding of azomethine nitrogen with metal ion. In the IR spectrum of ligands, stretching vibrations of ˃C=O group appeared at 1700-1720 cm-1 and SO2 group appeared at 1315-1350 cm-1 (asymmetric) & 1150-1247 cm-1 (symmetric) are not changed on complexation which indicates both groups (˃C=O & SO2) are not involved in coordination with metal. Two new bands appeared around at 519-555 cm-1 and 410 cm-1 due to ѵ M-N and ѵ M-Cl bonds confirm metal-ligand bonding, respectively.
Thermogravimetric Analysis (TGA)
The thermal decomposition analysis of synthesized complex C-3 was carried out under an inert nitrogen atmosphere (Fig.4). A weight loss of 8% was observed between 100-230oC due to loss of two chlorine atoms. Another thermal decomposition occurs between230-882oC with weight loss of 52%due to loss of remaining organic part. Total 60% weight loss was observed between 100-882oC. Heating rate was suitably controlled at 30oC min-1. Thus TGA curve indicates presence of two chlorine atoms in the tin complex and tetrahedral geometry has been suggested having 1:2 stiochiometric ratio of Sn(II) and ligand, respectively.
 |
Figure 4: (A) TGA curve (B) DTA curve for C-3 complex |
Differential Thermal Analysis (DTA)
In DTA curve of C-3 complex two downward peaks around 250-290oC and 450-470oC were observed which indicate reaction is endothermic in nature.
1H-NMR of azomethine nitrogen (–CH=N–) showed upfield shifting from δ 8.52-8.55 to 8.00-8.50 ppm in their tin complexes. This upfield shifting in complex indicate discharging of lone pair of nitrogen atom towards the metal ion. Methoxy proton appeared at δ 3.86-3.65 ppm as singlet, this indicates methoxy group is not involved in coordination with metal ion.
All aromatic protons shifted towards downfield region on complexation with the Sn metal ion. In C-1and C-3 complexes ortho protons of aryl shifted towards downfield region at δ 7.38 to 7.66 ppm and δ 6.78 to 7.01 ppm, respectively due to complexation with metal.
The proposed structures (Fig. 3.) of tin(II) complexes (C-1 to C-4) is in good agreement with the spectral data presented and reported literature45-46.
In13C-NMR spectra of C-3 complex, a downfield shifting was observed in the azomethine carbons from δ 158.14-162.39 ppm (free ligands) to δ 169.3-174.2 ppm (tin complexes) may be due to shifting of lone pair of nitrogen towards metal ion. Similarly all phenyl ring carbons shifted towards downfield region by 0.1-0.40 ppm.
Mass fragmentation pattern of all Schiff base ligands (L1-L4) and their complexes (C-1 to C-4) were recorded. LC-MS of Schiff base L3 and its tin complex C-3 were explained here. The molecular ion peaks at m/e 396.74 and 1010.96 attributed to the ligand & its complex, respectively. Mass spectra of the complex C-3 showed peaks at m/e 1010.96, 1012.25, 1014.58,1031.30 and 1016.85 corresponding to (M+), (M+2), (M+4), [C41H41Cl2N8O6S2Sn]+ and [C41H42Cl2N8O5S2Sn]+, respectively and confirms that complex contains two chlorine atoms.
Antibacterial Activity
Results of antibacterial activity indicate that the C-3 complex was found most active (MIC, 50 µg/mL) against the pathogen E. coli and C-1 complex was found with good activity (MIC, 55.6 µg/mL) against E. coli. Further C-2 was found more active and C-3, C-4 was found with good activity against gram negative P. aeruginosa. C-1, C-2 and C-3 complexes were found with good activity against S. aureus bacteria. Complex C-1 was found with better activity against S. pyogenus bacteria. L3 was found with better activity (MIC, 52 µg/mL) against the pathogen E. coli. Thus these results indicate that complexes exhibit better activity than the ligands.
Antifungal Activity
The antifungal activity of four Schiff base ligands (L1-L4) and their complexes (C-1 to C-4) were tested against fungal strains C. albicans, a. niger and a. clavatus. Complex C-3 exhibit better activity (MIC, 250 µg/mL) and complex C-1 was found with good activity (MIC, 500 µg/mL) against C. albicans. C-3 complex was found with better activity (MIC, 100 µg/mL) and C-1, C-2, C-4 complexes showed good activity against A. Niger. C-4 complex was found with good activity (MIC, 250 µg/mL) against A. clavatus bacteria. Greseofulvin drug was used against the bacteria. L3 and L4 showed with good activity against A. Niger and C. albicans (MIC, 250 µg/mL). These results indicate that the complexes show good activity than the ligands.
Antimalarial Activities
In vitro antimalarial activities of ligands (L1-L4) and their complexes (C-1 to C-4) were evaluated against parasite P. falciparum strain. Results of antimalarial activity were expressed asIC50value (Table-1). The smaller IC50value indicates higher the antimalarial activity. Chloroquine was used as control drug in this study. Complex C-3 was found more active against chloroquine (IC50, 0.04 µg/mL) and complexC-1 was found with good activity (IC50, 0.07 µg/mL) than other complexes. L3 showed with good activity against chloroquine (IC50, 0.10 µg/mL). These results indicate that complexes are more significant than the ligands.
Table 1: Result of antibacterial, antifungal and antimalarial activities of Schiff base ligands (L1-L4) and complexes (C-1 to C-4)
|
S. No. |
Compound |
Antibacterial activity (MIC, µg/mL) |
Antifungal activity (MIC, µg/mL) |
Antimalarial activity (MIC, µg/mL) |
|||||
|
E. Coli (MTCC 443) |
P. aeruginosa (MTCC 441) |
S. aureus (MTCC 96) |
S. pyogenus (MTCC 442) |
C. albicans (MTCC 227) |
A. Niger (MTCC 282) |
A. clavatus (MTCC 1323) |
P.falciparum |
||
|
1 |
L1 |
62.5 |
100 |
100 |
100 |
1000 |
500 |
1000 |
0.14 µg/mL |
|
2 |
L2 |
100 |
125 |
125 |
250 |
500 |
500 |
1000 |
1.36 µg/mL |
|
3 |
L3 |
52 |
125 |
61.3 |
100 |
500 |
250 |
500 |
0.10 µg/mL |
|
4 |
L4 |
125 |
250 |
100 |
125 |
250 |
250 |
500 |
0.19 µg/mL |
|
5 |
C-1 |
55.6 |
100 |
65.5 |
85.2 |
500 |
250 |
1000 |
0.07 µg/mL |
|
6 |
C-2 |
100 |
62.5 |
100 |
125 |
500 |
250 |
500 |
0.90 µg/mL |
|
7 |
C-3 |
50 |
100 |
51.5 |
100 |
250 |
100 |
500 |
0.04 µg/mL |
|
8 |
C-4 |
250 |
125 |
100 |
250 |
250 |
200 |
250 |
0.40 µg/mL |
|
|
Standard drugs |
25 |
25 |
50 |
50 |
500 |
100 |
100 |
0.020 µg/mL |
SAR studies
To look at the structure activity relationship (SAR) of sulpha drug Schiff base ligands, the variations were determined on one end of sulphonamide moiety with six membered two heteroatoms with alkyl groups. On the other hand azomethine linkage was responsible for biological activity. Among the four complexes (C-1 to C-4), complex C-3 was found to be best antibacterial, antifungal and antimalarial agent among the tested compounds due to sulphadiazine moiety and two methyl substituents.
Moreover, comparative study of biological activities of all the tested Schiff bases and their complexes revealed that Sn(II) complexes are more active than respective ligands.
Conclusion
In our present studies, sulpha drugs based Schiff bases and their Sn(II) complexes (C-1 to C-4) were prepared in MeTHF by conventional method and characterized by various techniques. Thus Schiff bases can bind with metal ion through N-atom and tetrahedral geometry is proposed. All the Schiff base ligands and their complexes were screened for antibacterial, antifungal and antimalarial activities. Complex having sulphadiazene moiety has presented good antibacterial, antifungal and antimalarial activity. It was concluded that Sn(II) complexes are more active as compare to respective ligands.
Acknowledgement
Kiran Meena is highly thankful to SAIF, Punjab University, Chandigarh for NMR & MASS analysis and for IR spectra, Department of Chemistry, M.L.S.U. Udaipur (Raj.), India. For biological studies authors are also grateful to Microcare Laboratory, Surat (Gujrat), India.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Sources
There are no Funding Source.
References
- Abbas, S.A.; Munir, M.; Fatima, A.;Naheed, S.; Ilyas, Z.; Electronic J. of Life Science., 2010, 1, 37-40.
- El-Mossalamy, E.H.; Al-Thabati, S.A.; Al-Nowalser, F.M.; Al-Sulami, Q.A.; Communications Faculty of Sciences University of Ankara Series B., 2005, 51, 21-30.
- Li, Y.; Yang, Z.S.; Zhang, H.; Cao, B.J.; Wang, F.D.; Bioorg.&Med. Chem., 2003, 11, 4363-4368.
CrossRef - Villar, R.; Encio, I.; Migliaccio, M.; Gil, M.G.; Martinez-Merino,V.; Bioorg.& Med. Chem.,2004, 12, 963- 968.
CrossRef - Panneerselvam, P.; Nair, R.R.; Vijayalakshmi, G.; Subramanian, E.H.; Sridhar, S.K.; Eur. J. Med.Chem., 2005, 40(2), 225-229.
CrossRef - Bhat, M.A.; Imran, M.; Khan, S.A.; Siddiqui,N.J.; Pharma. Sci., 2005, 67, 151-159.
- Wang, L.; Feng, Y.; Xue, J.; Li, Y. J.; Serbian Chem. Soc., 2008, 73, 1-6.
CrossRef - Wadher, S.J.; Puranik, M.P.; Karande, N.A.; Yeole, P.G.; Int. J. Pharm. Tech. Res., 2009, 1, 22-33.
CrossRef - Matela, G.; Anti-cancer agents in medicinal chemistry, 2020, 20, 1908-1917.
CrossRef - Zoubi, W.A.; Int. J. of. Org. Chem., 2013, 3, 73-95.
CrossRef - Prakash, A.; Adhikari, D.J.; Pharm. Tech. Res., 2011, 3-4, 1891-1896.
- Mostafa, A.A.; Al-Askar, A.A.; Almaary, K.S.; Dawoud, T.M.; Sholkamy, E.N.; Bakri, M.M.; Saudi J. of Biological Sciences, 2018, 25, 361-366.
CrossRef - Balouiri, M.; Sadiki, M.; Ibnsouda, S.K; J. of pharma. Analysis, 2016, 6, 71-79.
CrossRef - Silva, C.M.; Silva, D.L; Modolo, L.V.; Alves, R.B.; Reseude, M.A.; Martins, C.V.B.; Fatima, A.D.; J.of Adv. Res., 2011, 2, 1-8.
CrossRef - Chukwujekwu, J.C.; Staden, J.V.; Smith, P.; South African. J. of Botany, 2005, 71, 316-325.
CrossRef - Rudrapal, M.; Chetia, D.; Prakash, A.; Med. Chem. Res., 2013, 22, 3703-3711.
CrossRef - Chauhan, L.K.; Nimodia, K.; Ranawat, P.S.; Goswami, A.K.; Baroliya, P.K.; Oriental J.of Chem., 2020, 36(5), 855-862.
CrossRef - Regar, M.; Baroliya, P.K.; Patidar, A.; Dashora, R.; Mehta, A.; Chauhan, R.S.; Goswami, A.K.; Pharmaceutical Chemistry journal, 2016, 50, 5.
- Dayma, V.; Chopra, J.; Sharma, P.; Dwivedi, A.; Tripathi, I.P.; Bhargava, A.; Murugesan, V.; Goswami, A.K.; Baroliya, P.K.; Heliyon, 2020, 6, 4787.
- Patil, U.; Khan, A.; Nagarsekar, A.; Mandewale, M.; Yamgar, R.; Oriental J. of Chem., 2018, 34(6), 2796-2805.
CrossRef - (a) Supuran, C.T.; Casini, A.; Scozzafava, A.; Med. Res. Rev., 2003, 5, 535.(b) Scozzafava,A.; Owa,T.; Mastrolorenzo, A.; Supuran,C.T.; Curr. Med. Chem., 2003, 10,925.
CrossRef - Kleemann,A.; Engel,J.; Kutscher, B.; Reichert, D.; Eds. Pharmaceutical Substances, Syntheses, Patents, Applications., (Thieme, Stuttgart, 1999).
- Domagk, G. Chemotherapy of bacterial infections. Dtsch. Med. Wochensch., 1935, 61, 250-253.
CrossRef - Domagk G.A new class of disinfectants. Dtsch. Med. Wochensch., 1935, 61, 829-832.
CrossRef - Domagk G., Chemotherapy of bacterial infections. Angew. Chem., 1935, 48, 657-667.
CrossRef - Mahmood-ul,H.; J. Enzyme Inhibition& Med. Chem., 2004, 19, 263-267.
CrossRef - Singh,V.; Kaushik, N.K.; Singh,R.; Asian J. Research Chem., 2011, 4(3), 339-347.
- Brodowska, K.; Charucinska, E.L.; Chemik, 2014, 68, 129-134.
CrossRef - Davies ,A.G.; Smith ,P.J.; Wilkinson ,G.; Stone ,F.G.A.; Abel, E.W.; Comprehensive Org. Chem., 1982, 2, 519.
- Priyanka; Kumar, M.; Sharma, H.K.; Soni, S.; Oriental J. of Chem., 2020, 36(5), 871-878.
CrossRef - Shekhawat, V.S.; Varshney, S.; Varshney, A.K.; J. Indian Chem. Soc.,2017, 94, 21-28
- Armarego, W.L.F.; perin,D.D.4th ed., Butterworth Heinemann.1997.
- Barnabas, M.J.; Parambadath, S.; Nagappan, S.; Ha, C.S.; Heliyon, 2019, 5, 2405- 8440.
CrossRef - Hassan, M.; Nasr, S.M.; Razak, S.E.A.E.; Aziz, M.S.A.; Gamasy, S.M.E.; Arabian J.of.Chem., 2020, 13,7324-7337.
CrossRef - Clinical and Laboratory Standards Institute Methods for antimicrobial susceptibility testing of aerobic bacteria approved standard M07- A8, 9th edn. National Committee for Clinical Laboratory Standards, Wayne, 2008.
- Muller, J.H.; Hinton, J.; Proc. Soc. Exptl. Biol., 1941, 48, 330.
CrossRef - Downes, F.P.; Ito,K. Compendium of Methods for the Microbiological Examination ofFood 4th ed, APHA, Washington.2001.
CrossRef - Rieckmann, K.H.; Campbell, G.H.; Sax, L.J.; Mrema, J.E.; Lancet, 1978, 1, 221-223.
- Peters,W.; Richards, W.H.G.; Handbook of Experimental Pharmacology Springer-VerlagGermany,1984, 179-200.
- Trager, W.; Jensen, J.B.; Human Malaria Parasites in Continuous Cult. Sci., 1976, 193, 673-675.
CrossRef - Lambros, C.; Vanderberg, J.P.J.; Parasitol, 1979, 65, 418-420.
CrossRef - Singh, J.J.S.B. stain.; A review. Indian J. Malariology, 1956, 10, 117-129.
- Panjarathinam, R.; Text Book of Medical Parasitology, 2nd Ed., Orient Longman, 2007, 329-331.
- Abu-Khadra, A.S.; Afify, A.S.; Mohamed, A.; Farag, R.S.; Aboul-Enein, H.Y.; The Open Bioactive Compounds Journal, 2018, 6, 1-10.
- Kayed, S.F.; Farina, Y.; J. of Saudi Chem. Soc., 2020, 24, 236-243.
CrossRef - Bhatra, P.; Sharma, J.; Sharma, R.A,; Singh, Y.; Appl. Org. Chem., 2017, 31, 3639.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









