Activity of Laccase Enzyme Present in the Phenol-Contaminated Sediments of the Marilao-Meycauayan-Obando River System, Philippines
King Dave G. Martin1,2* , Maria Fatima T. Astrero1, Laurence Anthony N. Mallari1 and Roland M. Hipol1
, Maria Fatima T. Astrero1, Laurence Anthony N. Mallari1 and Roland M. Hipol1
1Department of Biology, College of Science, University of the Philippines – Baguio, Philippines.
2Department of Chemistry and Environmental Science, College of Arts and Sciences, Nueva Ecija University of Science and Technology, Cabanatuan City, Nueva Ecija, Philippines.
Corresponding Author E-mail: kgmartin@up.edu.ph
DOI : http://dx.doi.org/10.13005/ojc/370122
Article Received on : 22-Jan-2021
Article Accepted on :
Article Published : 24 Feb 2021
Laccases are enzymes produced by different microbes like bacteria and fungi. These enzymes are members of the family of oxidases and are capable of oxidizing phenolics into non-toxic forms. Sediments were collected from the Marilao-Meycauayan-Obando River System, specifically from the sampling area connected to leather tanneries, which directly dump their effluents into the river. This study aimed to determine the presence of laccase activity of sediments of Meycauayan River where effluents of leather factories and tanneries are directly dumped. Concentration of the phenolic compounds from five collection sites were measured. Collected phenol - contaminated sediments were tested for laccase activity using ABTS (2,2'-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid)). Laccase was extracted from the collected sediments and used for the degradation of phenol. Reduction of phenol concentration by the extracted laccase reached 79.82% to as high as90.84%with a starting phenol concentration of 27.5 mmol per sample. Three strains of phenol-degrading ligninolytic bacteria closely related to Lysinibacillus xylanilyticus, Lysinibacillus fusiformisss, and Lysinibacillus sphaericuswere identified to be present in the river’s sediment which could produce laccase and facilitate degradation of phenol.
KEYWORDS:Biodegradation; Enzyme; Laccase; Lysinibacillus; Marilao-Meycauayan-Obando River System; Phenol
Download this article as:| Copy the following to cite this article: Martin K. D. G, Astrero M. F. T, Mallari L. A. N, Hipol R. M. Activity of Laccase Enzyme Present in the Phenol-Contaminated Sediments of the Marilao-Meycauayan-Obando River System, Philippines Orient J Chem 2021;37(1). |
| Copy the following to cite this URL: Martin K. D. G, Astrero M. F. T, Mallari L. A. N, Hipol R. M. Activity of Laccase Enzyme Present in the Phenol-Contaminated Sediments of the Marilao-Meycauayan-Obando River System, Philippines Orient J Chem 2021;37(1). Available from: https://bit.ly/2ND96p8 |
Introduction
Water pollution has become an increasingly adverse environmental concern in the Philippines [1]. In 1996, the Department of Environment and Natural Resources (DENR) has confirmed that almost half of the rivers in the country are already polluted and unsuitable for beneficial use [2]. As a response, the government enacted the Republic Act (RA) No. 9275 known as the Philippine Clean Water Act of 2004. In line with this law is the designation of Water Quality Management Areas (WQMA) in consideration of the water quality, degree of pollution and the risks posed by the river [3]. Included in the first list of WQMA’s is the Marilao-Meycauayan-Obando River System (MMORS), one of the most polluted river systems in the country located in Bulacan where tanneries, leather factories and refineries are the most common industries [4].
One of the major pollutants of MMORS specifically Meycauayan River is the waste generated by tanneries and leather factories in the city. The accumulation of tannins [5] increases the concentration of phenols in the river. Phenols are aromatic organic compounds that has a hydroxyl group bound to an unsaturated aromatic hydrocarbon ring [6]. This compound was found to be a reproductive toxin as it increases the risk of abortion and low birth weight [7]. In addition, phenols may undergo several different reactions such as nitration, methylation, alkylation, or substitution with chlorine atoms which is harmful to living organisms. The toxicity of phenol is associated with the hydrophobicity of the individual compound, and the formation of free radicals. Hydrophobicity affects the solubility of phenol in a cell and consequently the possibility of interaction between the compound and specific cell and tissue structures [8]. The damaging effects of phenols and their derivatives entail acute toxicity, histopathological alterations, mutagenicity, and carcinogenicity.
Researchers have been conducting studies on bioremediation as an efficient means to eliminate toxic substances such as phenols. One of the most widely studied methods is the use of enzymes for their efficiency, high selectivity and nonhazardous reactions This method is also applied in the degradation of phenolic compounds. The most common is the use of laccase which is a glycoprotein that has four copper atoms distributed in its redox sites. Laccase acts on both phenolic and nonphenolic lignin-related compounds which contributes to their efficiency in bioremediation and wastewater treatment. Laccase only requires molecular oxygen to be able to catalyze oxidation reactions, contributing to its efficiency in xenobiotic degradation [9]. Besides its ability to degrade toxic substances, it can also reduce molecular oxygen to water [10] making it an interesting and important focus for research.
Although government and public efforts have been employed for the restoration and rehabilitation of the Marilao-Meycauayan-Obando River System, none of the cleanup drives specifically and specially aim to reduce the amount of harmful and toxic substances which have accumulated in the river. Laccase genes are capable of degrading phenolics into less-toxic forms. Through this study, an assessment of the degree of phenol contamination in the river was carried out. Sediments from the contaminated Meycauayan River was tested for its possible laccase activity. Moreover, a feasible way of reducing phenol through bioremediation was shown based on the results.
Materials and Methods
Field site visitation and collection of sediment
Sediments samples from the river were collected during dry season where water quality is better compared during wet season.Samples were collected in five random sampling points where most of the large-scale tanneries along the Meycauayan River dump their effluents using an improvised sediment collector. Sample 1 was collected in the middle of the banks, Samples 2 and 3 were collected from the opposite banks, Sample 4 was from a creek connected to the river where effluents of a large-scale tannery is directly dumped, and Sample 5 was from an area where effluents are directly dumped as well. Collected samples were preserved immediately prior to processing in the laboratory.
Sediment samples collected were stored im clean glass bottles and preserved using CuSO4 and H3PO4 to prevent biodegradation and contamination from other compounds [11] [12]. The collected sediment sampleswere preserved and kept at 4°C to avoid chemical degradation [7]since chemical changes occur in less than 24 hours if left unpreserved.
Quantification of phenolic compounds
Phenol concentration was quantified using Folin-Ciocalteu reagent basedfrom the methods used by Ainsworth and Gillespie [13]. Reagents used were 10% (vol/vol) Folin-Ciocalteu (F-C) reagent, 95% methanol solution and 700 nM Na2CO3. Gallic acid solution that served as the standardwas also prepared in varying concentration ranging from 0.0275 mM up to 2.75 mM that was used in constructing the standard curve. In different Eppendorfs tubes, 100 µL of sample, standard, and blank solutions were placed. To each of the Eppendorfs tubes, 200 µL of F-C reagent was addedand after the solutions were thoroughly mixed, 800 µL of 700 mM Na2CO3 was added and the solutions were incubated for 2 hours in room temperature for color development. After incubation, 200 µL of sample, blank, and standard solution were pipetted in a microplate and absorption in 765 nm was read. Since Folin-Ciocalteu (F-C) reagent only reacts with phenolic compounds, this assures that other compounds present in the samples will not interfere in the quantification of phenolic compounds.
Laccase extraction
Laccase was extracted from the soil samples following the report of McClaugherty and Linkins [14]. Laccase’s high specificity to phenol as substrate was taken as an advantage to ensure that other extracted enzymes did not interfere with the biodegradation process [15]. Fifty grams of soil was mixed with 300 mL of 50 nM acetate buffer at pH 5and was centrifuged for 10 minutes at 4,000 rpm, while maintaining temperature at 4° C. The supernatant was then filtered through vacuum filter with pore size of 40 micrometer. And the remaining pellet was suspended in 50 nM acetate buffer with pH 5 and 1M NaCl and was centrifuged. The supernatants acquired were combinedwhich is the crude laccase extracted.
Laccase assay
Activity of laccase enzyme was measured based from the methods of Airong Li and colleagues. [16] Four liters of working solution was prepared by mixing 3,800 mL of water with 200 mL of 1 M of sodium acetate stock solution with the pH adjusted to 5. Fresh solution of 2 mM ABTS was prepared using megapure water in 25-50 mL as required. As forthe soil sample preparation, 2.75 grams of sediment sample was added to 91 mL of 50mM sodium acetate buffer. The mixture was then homogenized using a blender and with the use of large orifice tips, 800 μL of the soil mixture was pipetted into the 96 well plates while continuously stirred to keep the slurry homogenized. Sodium acetate buffer and 200 µL of 2 mM ABTS were then added to designated well plates. The covered plates were incubated at 10°C for 6 hours and exactly 250 µL of the contents of the deep well plates were transferred to a different set of wells in an unused clear microplate. Readings were conducted using a microplate reader at 420 nm. Activity of the Laccase enzyme was measured using the following formula:
OD = (Sample + Substrate ABS) – (Sample Blank ABS) – (OD for ABTS+ Buffer)
Laccase Activity (μmol/h/g) = OD / (21.9 / μmol) (incubation time, h) (g sample / mL of sample homogenate)
Bioremediation of phenol by crude laccase extract
The ability of the extracted laccase to degrade phenol was analyzed using the methods done by Asadgol, et al. (2014) [17]. The highest concentration of the phenol detected from the river served as the basis for the initial concentration to be used in the bioremediation protocol. The reaction mixture was prepared by adding phenol to citrate buffer (20mM pH 5) in a 1:1 ratio. The extracted laccase was introduced to the mixture and was incubated at 35°C for 40 minutes. After incubation, the reaction mixtures were incubated at 35°C and samples were tested for remaining phenol content after one week. Phenol content was determined using Folin-Ciocalteu reagentby adding of 100 µL of laccase-treated phenol with 200 µL of F-C reagent and then by 800 µL of Na2CO3 followed by incubation at room temperature for 2 hours. The absorbance of the reaction mixture was determined at 765 nm. Standard curve was used to compute for the concentration of remaining phenol in mmol.
Identification of authocthonous bacteria
Collected sediment samples were homogenized and diluted. Ten grams sediments were dissolved in 90 ml of dH2O. This was followed by series of dilution until wanted concentration (10-6 dilution) was attained. [18] . Diluted samples were then plated to prepared nutrient agar plates and after a week, grown colonies of bacteria were isolated and purified following the standard microbial purification techniques.Nutrient agar plates containing the purified isolates were sent to Macrogen Inc. South Korea for DNA extraction, sequencing, and identification using universal rRNA primers. Identification was done using Basic Local Alignment Search Tool or BLAST [19].
Results and Discussion
Phenol contamination in Meycauayan River
Phenol contamination in Meycauayan River was assessed through spectrophotometric analysis. Figure 1 shows the phenol concentration in five different sampling points along the river. Sediment sample 4 was found to have the highest phenol concentration of 13.01 mmol L-1 as this was collected directly from the drainage site of a large-scale tannery along the river.
 |
Figure 1: Phenol concentration of sediment samples from Meycauyan River. |
The concentrations of phenolic compounds present in Meycauayan River ranged from 0.53 to 1.18 Gallic Acid Equivalent g-1 shown in Figure 1. The concentration of phenolic substance in the river reached up to 1,224 mg of phenol per liter of water, placing the river under IDLH or immediate danger to life or health concentration of phenol (19 mg m-3) by the United States Environmental Protection Agency (US EPA). This concentration of phenol in the river is toxic to humans and wildlife [20] hence, the National Institute of Occupational Safety and Health recommends exposure limit to ensure that a person can avoida condition that is likely to cause death.
All of the sampling sites in the river exceeded theUS EPA recommended phenol concentration in lakes and rivers which is 0.03 mg L-1. Sediment sample 4, which has the highest phenol concentration, was collected in a creek draining into the river and this creek is directly connected to a tannery. This suggests that the effluents from tanneriescontain high amount of phenolic by-products from the tanning process which contribute to the contamination in the river. Phenol is considered to be resistant to degradation[21], continuous release of phenol into the river system further increases the degree of contamination. As stated by Strong and Claus [22], high concentration of phenol in an environment can induce laccase formation in some fungi to degrade phenolic substance hence, there is a high possibility of laccase activity by indigenous organisms in the river [22].
Presence of laccase enzyme
The presence of laccase enzyme was tested through laccase assay. Shown in the Figure 2 is the computed laccase activity of sediments from Meycauayan River in µmol substratehour -1 g-1 which indicates the amount of laccase that oxidized 1 µmol of the substrate ABTS. Extracted laccase from sediment sample 2 showed the highest laccase activity of 0.05 µmol hour -1 g-1 of soil.
 |
Figure 2: Laccase activity of sediments collected from Meycauayan River. |
The laccase assay conducted was only used to detect and confirm the presence of laccase regardless of the rate of enzyme activity as sediment samples were directly used in the assay and no extraction was done. Thus, based on the results, even with very low laccase activity ranging from 0.000273 U to 0.000843 U, it can be concluded that laccase enzyme is present in the sediments.
The substrate used in the assay was ABTS which is highly specific for laccase enzyme. Upon oxidation of ABTS, it is readily converted to its cationic form resulting to a decrease in absorbance of ABTS at 420 nm indicating the presence of laccase. Laccase assay done directly with the sediment samples without prior laccase extraction is expected to yield lower activity compared with other literatures as observed in the results. This is because humic acids in sediments competitively inhibit the laccase enzyme [23]. To address this, hydrogen peroxide (H2O2) was added to enhance the activity of laccase in the soil by serving as an intermediate in the reduction of oxygen thus strengthening the results verifying the presence of laccase [24].
Theoretically, an increase in phenol concentration subsequently increases laccase activity of indigenous organisms in the environment. However, results do not show strong correlation between phenol concentration and laccase activity as illustrated in the Figure 3.
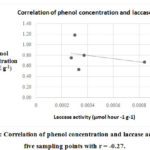 |
Figure 3: Correlation of phenol concentration and laccase activity in five sampling points with r = -0.27. |
The computed correlation coefficient (r) is -0.27 indicating a very weak to no significant downhill relationship between phenol concentration and laccase activity. Availability of oxygen in the sampling points vary and since oxygen is required for laccase enzyme activation, areas with low oxygen concentration also have reduced laccase activity. Moreover, phenol degradation by laccase activity may have commenced prior to phenol quantification resulting to inconsistent phenol concentration and laccase activity in the sampling points hence, there is very weak correlation between the two variables.
Biodegradation of phenol using crude laccase
The presence of laccase was confirmed through the laccase assay presented earlier. The ability of the extracted laccase to degrade phenol was tested and the results obtained are shown in Figure 4. Significant decrease in phenol concentration can be observed in all sediment samples after 40 minutes. However, there was little change in phenol concentration between the 40-minute degradation and the seven-day incubation time as observed in all the samples.
 |
Figure 4: Phenol concentration after treatment with crude laccase extract quantified after 40 minutes and 1 week. |
After analyzing the initial phenol concentration and the values after 40 minutes and one week on a Single Factor ANOVA at 0.01 significance level, results show that the P-value, 7.75 x 10-14 approaches zero, indicating that the values are unequivocally significantly different. The obvious drop in the concentration of phenol after the 40-minute mark ranging from 79.82% to 90.84 % degradation is an indication that laccase activity is present and ongoing. It can be deduced that laccases, which are oxidases, present in the crude samples have catalyzed the oxidation of phenol followed by the reduction of oxygen into water. Via the action of the unidentified microorganisms in the crude sample, the concentration of phenol decreases as water increases, thereby diluting the samples overtime. This was evident since the absorbance of the samples were significantly lower after 40 minutes and remained so even after a week. A slight increase in the phenol concentration in samples 4 and 5 between the 40-minute mark and one week of incubation was observed. However, this increase in phenol concentration is not and can therefore be accounted for by overreaction between Folin–Ciocalteu (FC) reagent and phenol which is important for color development [13]. Slightly prolonged incubation of phenol with F-C reagent may have caused this slight increase in phenol concentration.
Laccase is present in a wide variety of organisms, most notably in ligninolytic fungi and bacteria. The site from which the crude samples have been collected are surrounded by tanneries which dispense their waste into a neighboring Meycauayan river tributary. A large portion of these wastes are phenolic compounds from the commercial dyes used in making leather. The enzyme with the most promising results in terms of biodecolorization is laccase. The degradation of lignin, which is composed of cross-linked phenol polymers, stimulates laccase activity. The resulting low-molecular-weight metabolites from the degradation of lignin can mediate redox reactions, promoting the decolorization of azo dyes [25]. Numerous lignin-containing materials are used in tanning leather while azo dyes are the most common type of dye used in commercial leather manufacturing. These materials are then introduced into the river as waste. This surge in the phenolic compounds may have triggered the resident microorganisms in the river to produce an increased amount of laccase.
A river has the ability of self-purification through physical and chemical actions like dilution and the flow of water. The capacity of a river to heal itself is related to flow rate, sediment load, and organisms that can be found in the river. When the concentration of pollutants is beyond the abilty of the river to purify, the river becomes polluted [26]. As for the flow rate of Meycauayan river, the water is stagnant and no longer flowing and the apperance of water is dark and opaque indicating that the river is polluted. In contrary, the results from the test for bioremediation potential suggests that the river can potentially heal itself through the facilitation of its resident microorganisms. However, it is highly possible that the frequency of the dispensing of toxic wastes into the river waters outweighs the rate at which the laccase-producing microorganisms detoxify these wastes. Since these large-scale tanneries continuously drain their sewage into the river, its indigenous healing process becomes very slow, ultimately affecting the supposedly adequate resilience of the river.
Identification and autochthonous bacteria
Three different bacteria were isolated and identified to be related to Lysinibacillus species. These bacteria are Lysinibacillus xylanilyticus, Lysinibacillus fusiformisss, and Lysinibacillus sphaericus. Genus of Lysinibacillus are found to catabolically process different substrate such as ethanediol, organophosphorus pesticide Malathion, omeprazole, and dibenzothiophene. Lysinibacillus sp.was also isolated in the soils of textile industry area in lchalkarinji. In the study of Saratale and his colleagues, they were able to confirm laccase activity of isolated [27]. This supports that autochthonous bacteria in the river can degrade phenolic compounds. Laccase activity was also determined in the isolated Lysinibacillus sphaericus from the study conducted by Chantarasisri and his team (2017) [28]. Isolated bacteria from Meycauayan River with confirmed laccase activity contribute to the laccase activity of the sediments and biodegradation of phenolic substances. Presence of laccase producing bacteria is important for the treatment of wastewater from leather and dye industries [29].This also suggests that the river’s autochthonous microorganisms are contributing for the natural bioremediation of the river.
Conclusion
This study assessed the degree of phenol contamination in Meycauayan River and determined whether sediments from the river contain laccase enzyme. It was found that the degree of phenol contamination, reaching 13 mmol L-1 of water in the river is classified under immediate danger to life or health concentration by the US EPA. This concentration poses serious environmental and health risks and must be addressed by the local government. Despite the high amount of contamination in the river, laccase activity was confirmed and its efficiency in degrading phenol was found to be very high. Results revealed that within a span of 40 minutes, laccase from the sediment was already able to degrade up to 90.82% of the initial phenol. Three strains of phenol-degrading ligninolytic bacteria closely related to Lysinibacillus xylanilyticus, Lysinibacillus fusiformisss, and Lysinibacillus sphaericus were identified through Sanger sequencing. This suggests that the river can heal itself, but it is highly possible that the frequency of the dispensing of toxic wastes into the river waters outweighs the rate at which the laccase-producing microorganisms detoxify these wastes. It is recommended to have a follow up study on the optimization of methods on actual application of extracted laccase in river rehabilitation.
Acknowledgements
The authors would like to thank the Department of Biology in University of the Philippines – Baguio for funding this research. The authors would also like to thank the administration, faculty, and staff of the College of Arts and Sciences, Nueva Ecija University of Science and Technology for their support. Publication of the study was financially supported by the Nueva Ecija University of Science and Technology.
Conflict of Interest
The authors declare that there is no conflict of interest in this work with regards to publication
References
- Andrews, G.“Pepperdine Policy Review”, 2018, 10.
- Department of Environment and Natural Resources. “About Us | Water Quality Management Section.”, 2020. About Us | Water Quality Management Section (emb.gov.ph). [11/11/20].
- Malenab, M. C., Visco, E., Geges, D., Amparo, J. M., Torio, D., & Jimena, C. E. Analysis of the integrated water resource management in a water quality management area in the Philippines: The Case of Meycauayan-Marilao-Obando River System. Journal of Environmental Science and Management, 2016, 19(2).
- Department of Environment and Natural Resources. “What you should know about the Clean Water Act.”, 2004. http://emb.gov.ph/eeid/cwa- english.htm. [05/08/2020].
- Cornell University. Tannins: fascinating but sometimes dangerous molecules, 2015. http://poisonousplants. ansci.cornell.edu/ toxicagents/tannin.html. [09/08/2020].
- McMurry, J. Organic chemistry. Monterey, California: Brooks/Cole Pub. Co., 1984. 980-986
- University of California. Environmental Health and Safety: Phenols, 2017. https://ehs.ucsc.edu/lab-safety-manual/specialty-chemicals/ phenol.html. [05/08/2020].
- Michalowicz, J., Duda, W. Phenols – Sources and Toxicity, 2006. http://www.pjoes.com/pdf/16.3/347-362.pdf. [11/11/20].
- Viswanath, B., Rajesh, B., Janardhan, A., Kumar, A.P., Narasimba, G. Fungal Laccases and Their Applications in Bioremediation, 2014.
https://www.hindawi.com/journals/er/2014/163242/abs/[05/08/2020].
CrossRef - Madhavi, V., Lele, S. S. L. BioResources, 2009, 4(4), 1694-1717.
- Monitoring EP. Support Laboratory. Methods of chemical analysis of water and wastes. EPA-600/4-79-020. Cincinnati, OH: US Environmental Protection Agency, 1983.
- U.S. EPA. Methods for Collection, Storage and Manipulation of Sediments for Chemical and Toxicological Analyses: Technical Manual. Washington. U.S. Environmental Protection Agency, Office of Water, 2001.
- Ainsworth, E.A., Gillespie, K.M. Nature protocols, 2007, 2(4), 875-7.
CrossRef - McClaugherty, C., Linkins, A.E. Soil Biology and Biochemistry, 1990, 22(1), 29-33.
CrossRef - Atalla, M.M., Zeinab, H.K., Eman, R.H., Amani, A.Y., Abeer, A. A. E. A. Saudi Journal of Biological Sciences, 2013, 20(4), 373-381.
CrossRef - Li, A., Zhu, Y., Xu, L., Zhu, W.,Tian, X. African Journal of Biochemistry Research, 2008, 2(8), 181-183.
- Asadgol, Z., Forootanfar, H., Rezaei, S., Mahvi, A.H., Faramarzi, M.A. Journal of Environmental Health Science and Engineering, 2014, 12(1), 93.
CrossRef - Margesin, R., Schinner, F. Springer Science & Business Media, 2005, 5.
- Langaoen, A.F., Manzano, V.J., Requilman E.M., Tabardillo, J.M.,
Maningas, M.B., Calugay, R.J. Aquaculture, Aquarium, Conservation & Legislation, 2018, 11(2), 505-15. - Pradeep, N.V., Anupama, S., Navya, K., Shalini, H.N., Idris, M., Hampannavar, U.S. Applied Water Science, 2015, 5(2), 105-12.
CrossRef - Poi, G., Aburto-Medina, A., Mok, P.C., Ball, A.S., Shahsavari, E. Water, Air, & Soil Pollution, 2017, 228(3), 89.
CrossRef - Strong, P.J., Claus, H. Critical Reviews in Environmental Science and Technology, 2011, 41(4), 373-434.
CrossRef - Eichlerová, I., Šnajdr, J., Baldrian, P. Chemosphere. 2012, 88(10), 1154-60.
CrossRef - [24] Schlosser, D., Höfer, C. Applied and Environmental Microbiology, 2002, 68(7), 3514-3521.
CrossRef - Han, Y., Shi, L., Meng, J., Yu, H., Zhang, X. PLoS One, 2014, 9(10), e109786.
CrossRef - Tian, S., Wang, Z., Shang, H. Procedia Environmental Sciences, 2011, 11, 1328-33.
CrossRef - Saratale, R.G., Gandhi, S.S., Purankar, M.V., Kurade, M.B., Govindwar, S.P., Oh, S.E., Saratale, G.D. RGS, Journal of bioscience and bioengineering. 2013, 115(6), 658-67.
CrossRef - Chantarasiri, A., Boontanom, P., Nuiplot, N.O Aquaculture, Aquarium, Conservation & Legislation, 2017, 10(2), 20.
- Zhao, J., Wu, Q. X., Cheng, X. D., Su, T., Wang, X. H., Zhang, W. N., … Chen, Y. Frontiers of Chemical Science and Engineering, 2020, 1-16.

This work is licensed under a Creative Commons Attribution 4.0 International License.









