Quantifying Production of Quorum Sensing Regulated Pigments in Pseudomonas aeruginosa BIOTECH 1335
Chemistry and Environmental Science Department, College of Arts and Sciences, Nueva Ecija University of Science and Technology, Cabanatuan City, Nueva Ecija, 3100, Philippines.
Corresponding Author E-mail : krystelgrace_vergara@yahoo.com
DOI : http://dx.doi.org/10.13005/ojc/360519
Article Received on : 06 Aug 2020
Article Accepted on :
Article Published : 08 Sep 2020
One more method that can be used in the fight against communicable diseases is greatly important. Numerous pathogenic bacteria use intercellular signaling known as quorum sensing (QS) in defining virulence gene expression as well as gene regulatory mechanisms. Among the most promising sources of QSI agents are the ethnobotanicals. Extraction used 95% n-hexane in ethnobotanical leaves of A. triplinervis, B. pilosa, C. nocturnum, S. glabra, P. pentandrum, O. trinervis, D. elliptica, A. scholaris, A. adenophora, and Lipang Daga (no scientific name).
Extracts that were negative in the antibacterial testing proceeded to the QSI assay for pyocyanin production. The ten ethnobotanical extracts did not exhibit antibacterial activity against P. aeruginosa and were found to increase the pyocyanin production therefore the absence of QSI. However, all plant extracts can be used to increase the production of pyocyanin to accumulate more metabolites that are proven to have important biological and biotechnological applications.
KEYWORDS:Ethnobotanicals; Pyocyanin; Quorum sensing inhibition; Virulence factor
Download this article as:| Copy the following to cite this article: Padilla K. G. V and Martin K. D. G. Quantifying Production of Quorum Sensing Regulated Pigments in Pseudomonas aeruginosa BIOTECH 1335. Orient J Chem 2020;36(5). |
| Copy the following to cite this URL: Padilla K. G. V and Martin K. D. G. Quantifying Production of Quorum Sensing Regulated Pigments in Pseudomonas aeruginosa BIOTECH 1335. Orient J Chem 2020;36(5). Available from: https://bit.ly/3iecwc8 |
Introduction
Organisms have established numerous complicated ways to work together and adjust to the setting they occupy through natural evolution. For that reason, they must modify their phenotypes to respond in any diverse situations of external and/or internal pressure. Based on Van’t Padje et al. (2016), for an organism to thrive in the new environment they must alter their metabolism as well as other activities.1 Since the finding of antibiotics to combat infections, their utilization has been expansive. Microorganisms become accustomed to the misapplication of antimicrobial drugs resulted in their antibiotic-resistance. According to Van Hecke et al. (2017), within a short period of time antibiotics will not function properly and will become unmanageable in terms of communicable disease treatment and prevention, and also conventional contagions will be revived as of the main reason of mortality.2 In addition, the World Health Organization’s (2017) information’s are referring to the limitations of treatment selection to fight bacteria with resistance in antibiotics, and global synchronized action is vital to overwhelm this condition.3
According to the report of Starkey et al. (2014), the finding that numerous pathogenic bacteria employ cell to cell communication or termed as quorum sensing to control pathogenic effect and production of virulence factor assembly makes this process a striking focus for the strategy and progress of innovative curative agents.4 Any Gram type of the bacterium uses QS to organize collective actions as the purpose to assemble a large number of cells. The works of Lee and Zhang (2015) showed that the system of quorum sensing has been displayed to govern the creation of a collection of virulence factors outside the cells and the development of biofilm in numerous pathogenic bacteria from Pseudomonas aeruginosa is also included.5
One of the opportunistic pathogens found in human is P. aeruginosa that can specifically contaminate patients with acquired immune deficiency syndrome (AIDS), cancer, burn wounds, cystic fibrosis, cytotoxic drugs exposure, those with infections in the eye, blood, skin, and genitourinary tract infections as well as patients that undergo surgery and their immune system is compromised.6 Moreover, Girard and Bloemberg (2008) presented that this bacterium produces countless products that are released by the cells externally from where pyocyanin is included, and some are elastase, hydrogen cyanide, LasA protease, exotoxin A, alkaline protease, and rhamnolipids7. In order for P. aeruginosa to create and sustain the infection, it requires all these virulence factors that are released by the bacterium externally and are controlled by the QS system. Jakobsen (2011) recommended that deactivating the intercellular signaling mechanisms of a pathogen can end an important reduction in the production of these virulence proteins which regulate the disease mechanism of a bacterium.8 Therefore, the interference of the quorum sensing scheme provides a novel way to combat bacteria that are resistant to multiple antibiotics.
Indigenous peoples in the Philippines usually live in geographically remote areas and commercially valued natural resources are found concentrated in their areas, yet unexplored for their medicinal potential.9 Among these tribes is the Igorots of the Kalahan community called Ikalahans located in the Nueva Vizcaya, Philippines.10 Ethnobotanicals found in this area are used by the tribe for medical purposes, to control pests like rodents, and to paralyze fish for easy catching.11
There are no known studies about QSI potential of the ethnobotanicals in the Igorot community of Imugan, Nueva Vizcaya against pyocyanin production using n-hexane extracts which were the main target of the study, with great significance on the prevention and decrease in bacterial virulence.
Materials and Methods
Sample Collection
Plants included in the tests were pre-determined in an ethnobotanical survey12 through the permission of the council of elders of the Igorot community of Barangay Imugan, Santa Fe, Nueva Vizcaya. Ten plant samples were identified for testing and were collected from Mount Imanduyan located within the barangay. These were Bidens pilosa, Pittosporum pentandrum, Cestrum nocturnum, Oreocnide trinervis, Derris elliptica, Sarcandra glabra, Alstonia scholaris, Ayapana triplinervis, Lipang Daga (no known scientific name), and Ageratina adenophora. Leaves were collected by handpicking and were placed in clean, sealed plastic bags, and were transported to the laboratory for processing. Vegetative and reproductive parts of the specimens were collected in duplicates as required for obtaining correct identities. The authentication of the plants was carried out by an expert botanist from the National Museum of the Philippines in Manila.
Extraction Procedure with n-Hexane
Plant samples were cleaned with running tap water next rinse using distilled water and finally with 70% ethanol. Air-dried leaves were crushed to a fine powder using a blender9. Surplus pulverized leaves were kept in amber bottles or wrapped in clean plastic bags and stored in a cool dry place away from sunlight while waiting for use up to six months.9 Dried ground leaves with a weight of fifty (50) grams were soaked with a 500 ml of 95% n-hexane and kept in the Erlenmeyer or stoppered flask for 72 hours.9 Whatman No. 1 filter paper was used for filtration and was subject through a rotary evaporator.9 The filtrated extracts that were subject in rotary evaporator were kept in securely stoppered sterile amber bottles at refrigerated temperatures of 0-5℃13. Dimethyl Sulfoxide (DMSO) was used to dissolve crude n-hexane ethnobotanical extracts a final concentration of 20% plant extracts and 80% DMSO (100%).9
Sterilization followed by centrifugation of the crude extracts at 10,000 rpm for 30 minutes, then membrane filtration with a pore diameter of 0.45 μm.14 The extracts sterility was checked by inoculating 100 μl in brain heart infusion agar (BHIA) from time to time. The sterile extract was stored at 2-8℃ prior to use.14
Evaluation of Antibacterial Activity of Plant Extracts on Pseudomonas aeruginosa BIOTECH 1335 using Disk-Diffusion Assay
Pseudomonas aeruginosa BIOTECH 1335with a colony of three (3) to five (5) were full-grown for 16 to 18 hours in brain heart infusion agar (BHIA) at 35℃ and were shifted to sterile distilled water and McFarland 0.5 standard (~1.5 x 108 CFU/ml) was used to adjust the turbidity.9 Inoculation of Mueller Hinton Agar (MHA) plates was done by using a sterilized cotton swab moistened with the standardized culture. Sterile 6 mm paper discs (Sterile Blank Disc Hi-Media SD067) placed on sterile empty Petri plate with 20 μL of n-hexane plant extracts were pipetted and allowed to stand for a few minutes until excess liquid has flowed out. With the use of disinfected forceps, plant extracts in the discs were transferred onto 15-mm MHA plates equidistant to each other that were previously inoculated. Plates were prepared in triplicates. Norfloxacin (5 μg; Hi-Media SD184) served as the positive control while sterile distilled water served as a negative control.9 The presence of a clear or translucent zone of inhibition around the discs shows antibacterial activity.15 To eliminate an antibacterial effect that can decline in the production ofvirulence factor, which was vital for the precision of the succeeding assay, only ethnobotanical extracts lacking antibacterial activity were qualified for the pyocyanin production assay.
Evaluation of Quorum Sensing Inhibition in Pseudomonas aeruginosa BIOTECH 1335 Pyocyanin Production Assay
Overnight culture of Pseudomonas aeruginosa BIOTECH 1335grown in brain heart infusion broth (BHIB) was diluted to 0.06 OD at 600 nm using a UV-visible spectrophotometer (Beckman CoulterTM, DU®530 Life Science UV/Vis Spectrophotometer).16 Then, the 4.5 mL of P. aeruginosa culture was supplemented with 0.5 ml leave extract followed by incubation at 37°C for 24 hours.16 The treated culture was added with chloroform with the amount of 3 ml then mixing of the chloroform layer with 0.2 M HCl with a quantity of 1 ml. The extracted organic layer absorption was quantified with the used UV-vis spectrophotometer at 520 nm. For the blanks sample, sterile BHIB was used whereas P. aeruginosa culture of 4.5 ml with additional sterile distilled water of 0.5 ml was used as the control.
Statistical Analysis
Pyocyanin production is reduced in the treated culture at OD520nm. The significance of the OD measurements in pyocyanin production assay was determined using Mann-Whitney U, with a P<0.05 level of significance. Plant extracts with significantly lower OD measurements in comparison with the control meant the presence of QSI.
Results
Antibacterial Activity of Plant Extracts in Pseudomonas aeruginosa BIOTECH 1335
Antibacterial testing was performed in the n-hexane extracts of the ten (10) ethnobotanicals to select which extracts meet the requirements for the evaluation of QSI. Figure 1 shows the antibacterial activities of the ten n-hexane ethnobotanical extracts against P. aeruginosa BIOTECH 1335. All the ten n-hexane ethnobotanical extracts namely: A. triplinervis, B. pilosa, S. glabra, P. pentandrum, O. trinervis, C. nocturnum, D. elliptica, Lipang Daga (Unidentified), A. scholaris, and A. adenophora did not display antibacterial effect counter to the bacterium used in the study making them qualified for the succeeding virulence assays.
The results could be attributed to the polarity of the solvent used the n-hexane, a non-polar solvent extracted different set of phytochemicals but in a lesser amount that cannot suppress or inhibit the growth of the two test bacteria. It could also be accounted that the greater the phytochemical load, the greater the antibacterial activity of a plant extracts. Although there was no antibacterial effect, shifting the focus from the antibacterial to QSI properties of these ethnobotanical plants might lead to the discovery of new QS inhibitor compounds.
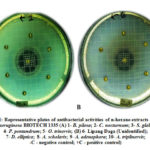 |
Figure 1: Representative plates of antibacterial activities of n-hexane extracts against P. aeruginosa BIOTECH 1335 |
Pyocyanin Production Assay
The mean OD520nm measurements of pyocyanin pigment production in the presence of ethnobotanical n-hexane extracts (mg/mL) which was diluted to 0.13 OD600nm is presented in Figure 2. All extracts did not result in any reduction in pyocyanin production in the test bacteria. Instead, extracts such as A. triplinervis, B. pilosa, S. glabra, P. pentandrum, O. trinervis, C. nocturnum, D. elliptica, Lipang Daga (Unidentified), and A. scholaris, resulted in higher pyocyanin output to significant levels compared to that of A. adenophora also with increased pigment production at 0.060 mg/mL, however, not significantly compared to the control which was at 0.027 mg/mL. As such, none of the ten n-hexane ethnobotanical extracts can be considered QSI to pyocyanin production. The increase may possibly be due to the nutritive effects of the extracts, promoting the expression of the virulence factor in P. aeruginosa although this has not been verified in this study.
 |
Figure 2: Mean optical density (OD) measurement of pyocyanin production in Pseudomonas aeruginosa BIOTECH 1335 with distilled water and n-hexane ethnobotanical extracts |
Discussion
Polar solvent has shown to recover more bioactive components from plants than nonpolar solvents hence, a greater phytochemical load.17,18 Many extracted phytochemicals from plants might not be highly effective as antibacterial agents.15 As an alternative, they hold properties against viruses or pathogenic bacteria, which are neither kill nor inhibit the growth of the bacteria and thus do not give the source or cause for the progress of bacteria which are resistant to various antibiotics.15 The reduction of gene expression for pathogenic bacteria and the release of virulence factors was mediated by these compounds by inhibiting communication between cells or intercellular signaling and some connected properties.19
Some studies have noted the absence of antibacterial effects in some bacteria of n-hexane extracts such as by Khatun et al. (2013) who reported that n-hexane and chloroform extracts displayed moderate inhibitoryeffect against most of the bacteria (including S. aureus and P. aeruginosa) in comparison to methanol or ethanol extracts.20 Ethyl acetate and n-hexane extracts showed no antibacterial activities against P. aeruginosa and S. typhi, respectively (Javid et al., 2015),21 whereas Singh et al. (2011), confirmed that ethanol, methanol, and water extracts of a plant, Tecoma stans can be effective against, E. coli, P. aeruginosa, S. aureus,and S. typhi.22 Biswas et al. (2013), likewise, indicated that only methanol and ethanol solvent extracts prepared from the leaves of Psidium guajava, showed inhibitory activities against bacteria (S. aureus and B. cereus), and no inhibitory results in n- hexane.23
Other studies, nevertheless, reported that some plant extracts have no QSI in pyocyanin production in the same bacterium such as the study of Tan et al. (2013), which reported that n-hexane and methanol extracts of Gnetum gnemon, and n-hexane extracts of Piper nigrum showed no significant inhibition against pyocyanin production.24 Ahmad et al. (2015), on the other hand, had similar results as this study had seven essential oil compounds increased pyocyanin production in P. aeruginosa.25 Panax pseudoginseng encouraged the making of pyocyanin in cultures that are older. The authors posited that there is an additional compound existing in the extract of ginseng, one compound is against swarming; the other compounds may disturb a dissimilar signaling trail in P. aeruginosa PAO1, inducing the synthesis of pyocyanin.26 These researches, on the other hand, specified that the volatiles of the plants are possible to either obstruct or encourage the cell to cell communication of the bacteria.
Although there were no QSI activities, all ethnobotanicals used in the study were able to enhance the production of pyocyanin. In other aspects of microbiology, this metabolite produces by P. aeruginosa have proven to use as dye decoloration,27 food colorants,28 antibacterial,28 antifungal,29 nematocidal29 and pesticide degradation,30 antioxidant, anticancer, plant growth promotion and is a promising biocontrol candidate,31 can be utilized for solving many agricultural27,28,30 and environmental issues.32
Extracts of A. triplinervis, B. pilosa, C. nocturnum, P. pentandrum, Lipang Daga, O. trinervis, A. scholaris, D. elliptica, A. adenophora, and S. glabra, do not exhibit antibacterial activities against P. aeruginosa. All ethnobotanical extracts show an increase in pyocyanin production of P. aeruginosa therefore the absence of QSI. Moreover, all plant extracts can be used to increase the production of pyocyanin therefore increasing the production of their metabolites that are proven to have important biological and biotechnological applications.
Acknowledgement
The authors recognize the consent, support, and help of the individuals from Imugan, Santa Fe, Nueva Vizcaya, Philippines. This research will not be possible without them so this is devoted to those people.
Conflict of Interests
The authors state that there is no conflict of interest.
References
- Van’t Padje, A.; Whiteside, M. D.; & Kiers, E. T. Current opinion in plant biology., 2016,32, 47-52.
CrossRef - Van Hecke, O.; Wang, K.; Lee, J. J.; Roberts, N. W.; & Butler, C. C. Clinical Infectious Diseases., 2017, 65(3), 371-382.
CrossRef - World Health Organization. No. WHO/EMP/IAU/2017.11., 2017., World Health Organization.
- Starkey, M.; Lepine, F.; Maura, D.; Bandyopadhaya, A.; Lesic, B.; He, J., … & Rahme, L. (2014). PLoS Pathogens., 2014, 10(8), e1004321.
CrossRef - Lee, J.; & Zhang, L. Protein & cell., 2015, 6(1), 26-41.
CrossRef - Ugurlu, A.; Yagci, A. K.; Ulusoy S.; Aksu, B; & Bosgelmez-Tinaz, G. Asian Pacific Journal of Tropical Biomedicine., 2016,6(8), 698-701.
CrossRef - Girard, G.; & Bloemberg, G. V. Future Microbiology., 2008, 3(1): 97-106.
CrossRef - Jakobsen, T. H.; van Gennip, M.; Christensen, L. D.; Bjarnsholt, T.; & Givskov, M. Methods in Molecular Biology., 2011, 692: 253-63.
CrossRef - The International Work Group for Indigenous Affairs (IWGIA). The Indigenous World., 2013, ISBN: 978-87-92786-33-3.
- Padilla, K. G. V.; Jacinto, W. R.; & Judan-Cruz, K. G. Advances in Bioresearch., 2018, 9 (6), 07-13.
- Villamor, G.; & Pindog, M. Bogor, Indonesia, World Agroforestry Centre -ICRAF, SEA Regional Office., 2008, 67.
- Undan, J. R.; Cruz, K. J.; Gandalera, E. E.; Abella, E. A.; David, E. S.; Valentino, M. J. G.; & Reyes, R. G. Central Luzon State University, Science City of Muñoz, Nueva Ecija., 2014, (Unpublished).
- Rezaei, A.; Oyong, G. G.; Borja, V. B.; Inoue, M.; Abe, T.; Tamamura, R.; … & Buery, R. R. Journal of Hard Tissue Biology., 2011, 20(2), 115-124.
CrossRef - Srisawat, S. Doctoral dissertation, Prince of Songkla University., 2007.
- Chenia, H. Y. , 2013, 13(3), 2802-2817.
CrossRef - Barrogo, K. N.; Jacinto, W. R.; & Judan Cruz, K. G. International Journal of Biosciences., 2018, 13(4), 173-182.
CrossRef - Adebayo, E. A.; Ishola, O. R.; Taiwo, O. S.; Majolagbe, O. N.; & Adekeye, B. T. African Journal of Plant Science., 2009, 3(12), 283-287.
- Wang, G. X.; Zhou, Z.; Jiang, D. X.; Han, J.; Wang, J. F.; Zhao, L. W.; & Li, J. Veterinary Parasitology., 2010, 171(3-4), 305-313.
CrossRef - Packiavathy, I. A. S. V.; Agilandeswari, P.; Musthafa, K. S.; Pandian, S. K.; & Ravi, A. V. Food Research International., 2012, 45(1), 85-92.
CrossRef - Khatun, M. A.; Alam, S. M. N.; & Hasan, M. N. Journal of Innovation and Development Strategy., 2013, 7(3): 7-9.
- Javid, T.; Adnan, M.; Tariq, A.; Akhtar, B.; Ullah, R.; & Abd El-Salam, N. M. African Journal of Traditional, Complementary and Alternative Medicines., 2015, 12(3), 91-96.
CrossRef - Singh, V.; Kumar, L.; Chakraborthy, G. S.; & Mazumder, A. Journal of Advances in Pharmacy & Healthcare Research, 2011, 1(3), 75-81.
- Biswas, B.; Rogers, K.; McLaughlin, F.; Daniels, D.; & Yadav, A. International journal of microbiology, 2013, 746165,7.
CrossRef - Tan, L. Y.; Yin, W. F.; & Chan, K. G. , 2013, 13(3), 3975-3985.
CrossRef - Ahmad, A.; Viljoen, A. M.; & Chenia, H. Y. Letters in applied microbiology., 2015, 60(1), 8-19.
CrossRef - Yeo, S. S. M.; & Tham, F. Y. Malaysian Journal of Microbiology., 2012, 8(1), 11-20.
- Bhatt, N.; Patel, K. C.; Keharia, H.; & Madamwar, D. Journal of Basic Microbiology: An International Journal on Biochemistry, Physiology, Genetics, Morphology, and Ecology of Microorganisms., 2005, 45(6), 407-418.
CrossRef - Saha, S.; Thavasi, R.; & Jayalakshmi, S. (2008). Research Journal of Microbiology., 2008, 3(3), 122-8.
- Kavitha, K.; Mathiyazhagan, S.; Sendhilvel, V.; Nakkeeran, S.; Chandrasekar, G. O. P. A. L.; & Dilantha Fernando, W. G. Archives of Phytopathology and Plant Protection., 2005, 38(1), 69-76.
CrossRef - Satapute, P.; & Kaliwal, B. Annals of Microbiology., 2016, 66(4), 1355-1365.
CrossRef - Babu, A. N.; Jogaiah, S.; Ito, S. I.; Nagaraj, A. K.; & Tran, L. S. P. Plant Science., 2015, 231, 62-73.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.










