Synthesis, Structural Characterization, Molecular Docking of Thiophene Derived Schiff Base from 4- Amino Indane and Antibacterial, DNA Binding Activities of its Nickel and Cobalt Complexes
K. Susmitha*, Manan Bhargavi, D. Sumalatha and Alina Jyothi Joseph
Department of Chemistry, St. Francis College for Women, Street No: 6, Umanagar, Begumpet, Hyderabad, Telangana, 500016, India
Corresponding Author E-mail: susmithakasula@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/360404
Article Received on : 05 Jun 2020
Article Accepted on : 06-07-2020
Article Published : 10 Jul 2020
Novel Schiff base is synthesized from 1:1condensation of thiophene 2- carboxaldehyde and 4-Amino indane. Two solid transition metal complexes (Ni and Co) are synthesized with this ligand. The structure of ligand (2-THIO- L) and its complexes are confirmed by various spectral characterization methods. 2-THIO- L and its metal complexes are tested for antibacterial activity against Staphylococcus aureus and Escherichia coli. Both show more activity against gram +ve than gram –ve bacteria. Molecular docking studies of ligand (2-THIO- L) against MAP2K6 indicated that they are potential inhibitors of carcinogenic receptors. DNA binding interactions of the Nickel and Cobalt complexes with CT-DNA in tris hydrochloride buffer at PH-7.2 are investigated using UV- Visible spectroscopy. The binding constants are in the range of 105 M-1 showed hypochromism which indicate good binding affinity towards CT–DNA.
KEYWORDS:2-THIO- L; 4-AMD; CT – DNA; DNA Binding; MAP2K6; Metal Complexes; Molecular Docking
Download this article as:| Copy the following to cite this article: Susmitha K, Bhargavi M, Sumalatha D, Joseph A. J. Synthesis, Structural Characterization, Molecular Docking of Thiophene Derived Schiff Base from 4- Amino Indane and Antibacterial, DNA Binding Activities of its Nickel and Cobalt Complexes. Orient J Chem 2020;36(4). |
| Copy the following to cite this URL: Susmitha K, Bhargavi M, Sumalatha D, Joseph A. J. Synthesis, Structural Characterization, Molecular Docking of Thiophene Derived Schiff Base from 4- Amino Indane and Antibacterial, DNA Binding Activities of its Nickel and Cobalt Complexes. Orient J Chem 2020;36(4). Available from: https://bit.ly/2BMuW3V |
Introduction
Schiff bases are the privileged ligands which gained importance due to their stability and flexibility.1 They are universal because of their easy synthesis, extensive applications and the ease of different structural modifications.2,3 The azomethine group is responsible for various antifungal, herbicidal, antibacterial and analytical activities.4,5 Aminoindanes form an important class of novel psychoactive drugs used for psychotherapy.6,7 They are used for ecstasy and higher dosages lead to serotonin syndrome. Schiff bases containing 4- Aminoindane became significant in medical and pharmaceutical fields due to its wide spectrum of activities like anticancer agents, bronchodilators, anticonvulsants and anti-HIV agents.8-11 Schiff base synthesis and its metal complexes containing 4-aminoindane with Nitro phenol are reported.12,13 Our published research paper reported the synthesis of Schiff base containing 4-Aminoindane with 2- hydroxyl benzaldehyde.14
MAP Kinases belongs to protein kinases that catalyze transfer of phosphate group from ATP to amino acid residues. Non phosphorylated MAP2K6 crystal structure was refined with 3VN9 in a putative auto inhibition form. Human Non- phosphorylated MAP2K6 crystal structure was binded with an Adenosine Tri Phosphate analogue and measured at 2.6 Aº resolution.15 Mitogen-activated protein Kinase kinase 6 (MAP2K6) belongs to dual specificity protein kinase family.16Receptor tyrosine kinases activate a number signaling pathways upon ligation of receptors in which mitogen activated pathway was one of them.17
Apart from this, small organic molecule like Schiff base, complexes with metal cations and interact effectively with macromolecules like DNA, RNA and peptides. As a result DNA binding studies of metal complexes gained significant interest over the past decades. Many compounds by binding to DNA obstruct the multiplication of tumour cells which forms the basis for development of novel anticancer agents. Very little information is existing so far on aminoindanes. Their increasing importance in medicinal research, has motivated us to carry out synthesis of novel Schiff base (2-THIO-L) derived from 1:1 condensation of 4-Amino indane (Structure-1) and thiophene 2 –carboxaldehyde (2-THIO) and also to perform Molecular docking. As continuation synthesis, characterization of Co (III) and Ni (II) metal complexes of (2-THIO-L), their antibacterial activity and the binding affinity with calf thymus DNA are also studied.

Materials and Methods
Materials
All chemicals used were of analytical purity. 4- Aminoindane (4-AMD), Thiophene 2–carboxaldehyde (2-THIO), Triethyl amine, Nickel (II) chloride hexahydrate, Cobalt(II)chloride hexahydrate, Dimethylsulphoxide (DMSO) and Calf thymus DNA were obtained from Aldrich Chemical Company Ltd. Solvents such as ethanol, Acetic acid were purified by standard methods. Electronic spectra were recorded with Systronics 2201 Double beam UV visible spectrometer ranging 200 – 1100 nm. Infrared spectra were measured using KBr pellets on Brucker Fourier Transformation spectrophotometer ranging from 400 to 4000 cm-1. 1H NMR spectra were determined using CDCl3 solvent, at Brucker 400MHZ spectrometer and developed by Mestrec 23 Software. The mass spectra were determined using ESI and LCMS (APCI Probe). The petriplates for antibacterial activity were photographed by using a Smartphone camera (Samsung galaxyM30). For docking studies biological databases like RCSB protein data bank and softwares like Autodock vina, Mercury 3.7 server have been used.
Synthesis of Schiff Base
Dissolve (1.13×10-2 moles) of 4-aminoindane in 20mL of 99% pure ethanol and (1.13×10-2 moles) of thiophene 2 –carboxaldehyde solution in 20mL of 99% pure ethanol separately. Mix both the solutions in which a few drops of acetic acid was added and the mixture was refluxed for about 4 hours. On cooling the compound was collected in ice as orange-yellow product which was filtered and recrystallized twice with ethanol. Synthesized Schiff base was obtained as golden yellow crystals, yield 80%, M.P. 148-1500C. (Scheme – 1)
 |
Scheme 1: Preparation of Schiff base ligand with 4- amino indane and thiophene 2 – carboxaldehyde |
Synthesis of Metal Complexes
The various metal complexes of Co (III) and Ni (II) were prepared by refluxing the corresponding 0.3mmol of the metal (II) chloride solutions with 0.60 mmol of the Schiff base solution (2-THIO-L), along with 0.06mmol of triethylamine in 30mL ethanol for about 4 hours. This resulted in green colored solution for Nickel complex and violet colored solution for Cobalt complex. The solutions were evaporated and colored precipitates were washed with ethanol and dried over anhydrous CaCl2. Yield 70 -75%, M.P.-150-3000C.
Molecular Docking
Molecular docking helps in studying binding interactions between receptor and ligand. With these interactions one can predict the affinity of binding and activity of ligands to the macromolecular protein receptors.
3VN9 crystal structure of Homo sapiens was downloaded from the RCSB PDB Bank. All the hetero atoms were removed from crystal structure of 3VN9.PDB in order to make receptor free from any ligand molecule before docking.
Extracellular signal regulated kinases, C-jun N terminal kinases and p38 kinases belong to mammalian MAPK family. These kinases undergo signaling pathway during cell proliferation. MAP2K6 was one of the important p38/ MAPK signal pathway.18 Under inflammatory and stress conditions MAP2K6 phosporylate and activates p38/ MAPK signal pathway (Fig.1). MAP2K6 was associated with various physiological and pathological processes such as cell proliferation, progression, division and inflammatory reactions. MAP2K6 is over expressed in many cancers such as esophageal, stomach and colon. MAP2K6 over expression was not only in these cancers but also in kidney, intestine, and lung cancers indicating its significance in human cancers and act as biomarker to predict or to diagnose the cancers.19 As 4-aminoindane derivatives possess anticancer properties, MAP2K6 was selected to carry out docking studies with the synthesized ligand (2- THIO- L)
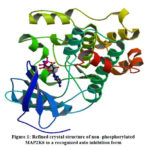 |
Figure 1: Refined crystal structure of non -phosphorylated MAP2K6 in a recognized auto inhibition form |
Protein Preparation
Protein was prepared and saved by adding polar hydrogens, salvation parameters and charges.
Ligand Preparation
The ligand (2- THIO- L) molecule was drawn in chemsketch, which was optimized and saved as MDL molfile. The saved ligand molecule was opened in Mercury 3.7 server2 and saved as .mol2 for docking purpose. In Auto Dock tool ligand preparation has been carried out by detecting roots, choosing torsions and by setting the number of torsions in Torsion Tree and saved in pdbqt format. 20-22
Grid Generation
Grid box was generated for the macromolecule with prepared ligand by taking active site amino acid residues from PDB sum and assigned x (-0.442), y (-0.558) and z (0.070) co-ordinates from Mercury 3.7 server2 and saved the molecule as GPF (grid parameter file). After running out auto grid GLG file had been generated this implies the successful generation of grid.
During the docking process DPF file was created and after the process DLG file had been created with which the process was completed. We analyzed the results in DLG file.
Antibacterial Activity
The antibacterial activity of the synthesized ligand (2-THIO-L) and its Nickel, Cobalt complexes were investigated against one G (+) i.e. Staphylococcus aureus and one G (-) i.e. Escherichia coli bacterial strains using agar diffusion process. The agar medium with concentration (2grams / 100 mL) was autoclaved and then placed in Petri plates for solidification. Three wells (9mm) were bored in the agar medium and 0.06μL of ligand solution and metal complexes with concentration of 5mg/mL in ethanol were placed in separate labelled wells. Incubation was carried out at 37oC for a day and zone of inhibition (in mm) was calculated against control for antibacterial assay.23
Electronic Absorption Spectra
Metal ion–DNA interactions can be explored using the experimental method named Ultra Violet absorption spectroscopy. When a metal complex gets inserted into a macromolecule there will be a deviation in its electronic absorption spectrum. The ligand field transition of the metal complex is disturbed when a base binds with it and results in red shift and blue shift, if the binding is intercalative. This is due to the strong stacking interaction between an aromatic chromophore and the base pairs of DNA. The degree of Hypochromism is directly proportional to the percentage of intercalation.24, 25
UV Absorption titration was carried out using constant concentrations of the complexes with increasing volume (0-100μl) of CT-DNA. While conducting the titration, fixed volume of Calf Thymus DNA was added to complex as well as blank in order to remove interference with CT-DNA. The equation used for calculation of intrinsic binding constant is given as
[DNA]/(εa-εf)=[DNA]/(εb-εf)+1/Kb(εb-εf) (1)
The concentration of DNA used is represented as [DNA], εa, εf and εb represents absorption coefficients, which relate to Aobs./[complex], where Aobs. is the extinction coefficient of the free complex and [complex] that of the DNA bound complex. A plot of [DNA]/ [εa-εf] against [DNA] provided a slope of 1/ (εb-εf) and intercept was calculated as 1/Kb (εb-εf). The ratio of the slope to the intercept gives the value for Kb.26
Results and Discussion
Physical Properties
The colour, yield and melting point of newly synthesized ligand and its metal complexes are summarised in Table 1.
Table 1: Physical properties of Ligand and its Metal Complexes
|
Compound |
Empirical formula |
Colour |
Yield |
M.P |
|
Ligand |
C14H13NS |
Yellow |
80% |
148-1500C |
|
Nickel Complex |
C28H28N2NiOS2Cl2 |
Green |
75% |
150-3000C |
|
Cobalt Complex |
C28H26CoN2S2Cl3 |
Violet |
73% |
150-3000C |
Spectral Interpretation of 2-THIO-L
The structure of 2-THIO –L was interpreted by UV, Infrared, H1NMR and Mass spectral analysis.
UV- Visible Absorption Spectra
The UV –Visible absorption spectra of newly synthesized ligand and its Nickel, Cobalt metal complexes were observed in DMSO solvent from 200 to1000 nm (Fig.2). The UV spectrum of ligand (2-THIO –L) displayed two absorption peaks at 280 nm and 360 nm as a result of π→π* and n-π* transitions. The UV spectrum of Ni (II) complex exhibited three bands at 250 nm, 340 nm and 420 nm. The first two absorption bands are because of ligand to metal charge – transfer transitions. The last absorption band is a result of d-d transitions and is given as 3A2g (F) → 3 T1g (P) . Co (III) complex showed two absorption bands around 230 nm and 280nm due to 5T2g→ 5Eg. The above observations suggest an octahedral geometry around Ni (II) and Co (III) ions in their complexes.27
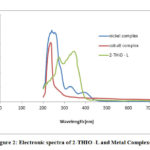 |
Figure 2: Electronic spectra of 2-THIO -L and Metal Complexes |
Infrared Spectral Interpretation
Infrared spectra of newly synthesized ligand displayed the following characteristic bands; sp3 C-H stretching (3013 cm-1), C=CH Aromatic stretching (2945-2970 cm-1) , azomethine HC=N stretching (1620-1629 cm-1), C-C bond stretching in aromatic ring (1593-1560 cm-1) and C-N stretching in aromatic amines (1215 cm-1 ) respectively(Fig.3).
 |
Figure 3: IR Spectra of the ligand |
1H NMR Spectra
The 1HNMR of 2- THIO- L in DMSO-d6 displayed a peak at 8.6 δ was due to azomethine proton and the aromatic protons resonances in the range of 6.7 to 7.7δ. The aliphatic protons of amino indane ring resonate in the range of 1.5-3.3δ (Fig.4).
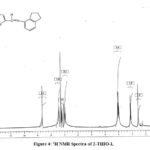 |
Figure 4: 1H NMR Spectra of 2-THIO-L |
Mass spectral Interpretation
The ESI- MASS spectra of 2-THIO-L showed a base peak at m/z = 238.18 whose expected mass was 227.08(Fig.5).
 |
Figure 5: ESI-Mass Spectra of 2-THIO-L |
Molecular Docking Results
Auto-Dock 1.5.6 docking software was used for Auto grid and Auto dock processes. The docking was used to define the orientation of inhibitors bound in the active site of receptors.
There were 10 conformers generated from each molecule in which best conformer is searched with Lamarckian Genetic Algorithm (LGA). Docking studies of the synthesized ligand was evaluated against MAP2K6 which was responsible for esophageal, stomach and colon cancer.
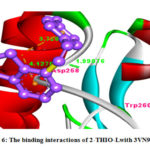 |
Figure 6: The binding interactions of 2-THIO-Lwith 3VN9 protein. |
The 2-THIO-L was showing binding interaction with Asp-258 and Trp-20(Fig.6). It was also showing negative binding energy value (-2.87) with 3VN9 protein which suggest that they act as potent inhibitors for the MAP2K6 protein to suppress cancer. From above discussion it was understood that the synthesized ligand possessed important inhibiting property for cancer causing receptors and act as anticanceragents.
Spectral Characterization of Metal Complexes
The Infrared, proton NMR and LCMS data of the novel complexes were summarized as below. The Infrared spectra of the complexes and that of 2-THIO-L were matched to decide the binding sites in the complexes. The 2-THIO-L, has an uncoordinated C=N stretching band at 1620cm-1 which is moved to 1616 cm-1 in Cobalt(III), 1499cm-1 in Nickel (II) complexes. This confirms that azomethine group is involved in bonding to metal ions. Stretching bands due to (M—X) at 810-550 cm–1 and (M—N), (M-S) at 500 – 315cm–1; in metal complexes was the further proof of coordination (Fig.7a, 7b).28
 |
Figure 7a: Infrared spectrum of Cobalt (III) metal complex |
 |
Figure 7b: Infrared Spectrum of Nickel (II) metal complex. |
Mass Spectra
The LC-MS data of the complexes of Nickel and Cobalt displayed the molecular ion peak at m/z 601 (M+) and m/z 619 (M+) respectively as shown in Fig. 8a and 8b.29
 |
Figure 8a: Mass spectrum of Nickel (II) – 2-THIO-L complex |
 |
Figure 8b: Mass spectrum of Cobalt (III) – 2-THIO-L complex |
Geometry of the Metal Complexes
From LCMS and IR spectral data the predictable geometry of Nickel and Cobalt metal complexes of 2-THIO- L were found to be octahedral (Fig. 9).
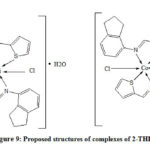 |
Figure 9: Proposed structures of complexes of 2-THIO-L |
Results of Antibacterial Activity
The antibacterial results of 2-THIO-L and its metal complexes are found to be active against Staphylococcus aureus (Gram +ve) but did not show any activity against Escherichia coli (Gram -ve) bacterial species (Fig.10a, 10b). Results confirm that the novel Schiff base and its complexes showed moderate activity.
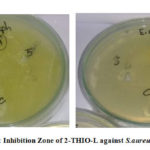 |
Figure 10a: Inhibition Zone of 2-THIO-L against S.aureus and E.coli |
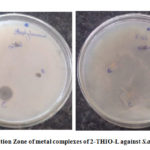 |
Figure 10b: Inhibition Zone of metal complexes of 2-THIO-L against S.aureus and E.coli |
Results of DNA Binding Studies
Metal complexes having planar aromatic groups gets inserted into the grooves of CT-DNA through non covalent interactions which leads to red shift and blue shift. The interaction of the complexes with Calf Thymus-DNA was observed by measuring the changes in the Ultra Violet absorbance of the complexes with the increase in the amount 0-100μl of CT-DNA (Fig.11a, 11b). Adding soluble form of calf thymus DNA to metal complexes in 5mM TrisHCl buffer at pH 7.2 showed hypochromism . The calculated Kb values for metal complexes are 1.22 x 105 M-1, and 1.66 x 105 M-1 respectively. So, it was evident that chromophoric groups of the synthesized metal complexes were involved in intercalative mode of interactions with DNA base pairs.
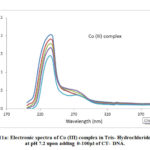 |
Figure 11a: Electronic spectra of Co (III) complex in Tris- Hydrochloride |
 |
Figure 11b: Electronic spectra of Ni (II) complex in Tris-Hydrochloride |
Conclusion
The 4- Amino indane derived Schiff base ligand (2-THIO-L) and its Nickel, Cobalt complexes have been prepared and analyzed spectroscopically. Molecular docking studies revealed that the synthesized ligand was potential inhibitor of cancer causing receptor. The results which we obtained from Auto-Dock were precise and computationally fit for docking of ligand to its specific receptor. From binding constant values Kb, it is observed that interactions between metal complexes and Calf Thymus DNA involves intercalation and results in Hypochromism. Molecular Docking and antibacterial studies reveal that derivatives of 4- Amino Indane acts more effectively as anticancer agents.
Acknowledgement
Authors are thankful to St. Francis College for Women Departments of Chemistry and Microbiology, for providing lab facilities to carry out the Research work. We also express our gratitude to Central Facilities for Research and Development (CFRD), Osmania University, Hyderabad and Sapala Organics Private Limited, Mallapur for the Instrumentation facilities to carry out Spectral characterization.
Conflict of Interest
Authors declare no conflict of interest.
References
-
- Djedouani, A.; Bendaas, ; Boufas, S.; Allain, M.; Bouet, G.; Khan, M. Acta. Cryst. 2007, E63, 1271-1273.
CrossRef - Hafeez, Ullah.; Feroza, H.W.; Muhammad, G.; Syed, A.T.; Sadia, A.W. J .Biochem. 2012, 37 (4), 386-391.
- Dhar, D. N.; Taploo, C. L. JSIR. 1982, 41( 8),501–506.
CrossRef - Yang, L.; Tai, X. S.; Qin, W. W.; Liu, W. S.; Tan, M. Y. Anal. Sci. 2004, 20, 357-361.
CrossRef - Jarrahpour, A.; Motamedifar, M.; Pakshir, K.; Hadi, N.; Zarei, M. Molecules. 2004, 9, 815-824.
CrossRef - Nikola, P.; Rachel, R. H.; Tomas, P. Psychiatry. 2017, 8, 236.
CrossRef - Linda, D. S.; Anna, R.; York, Schramm.; Marius, C. H.; Matthias, E. L. Pharmacol. 2014, 88(2), 237- 244.
CrossRef - Stewart, J. M.; Lajos, G.; Whalley, E. T. PCT Int. Appl. 1996, 1-21.
CrossRef - Gademann, K.; Chavez, D. E.; Jacobsen, E. N. Chemie. 2002, 41(16), 3059-3061.
CrossRef - KoteswaraRao, N. S. R. R. M. M.; Ram Reddy, M. G. Biometal. 1990, 3, 19-23.
CrossRef - Heinzelmann, R. V.; Kolloff, H. G.; Hunter, J. H.; Upjohn, C.; Kalamazoo, M.I. Am. Chem. Soc. 1948, 70, 1386-1390.
CrossRef - Osowole, A. A.; Daramola, A. O. Elixir Appl. Chem. 2012, 47, 8662-8666.
- Osowole, A.; Ingo Ott.; Oladunni, M. O. int. j. inorg. chem. 2012, 1-
- Arthi, S.; Sumalatha, D.; Susmitha, K.; Bhargavi, M.; Alina Jyothi, J. Asian J. Chem. 2020, 2(3), 539– 542.
CrossRef - Takashi, ; Takayoshi, Kinoshita.; Hitomi, Matsuzaka.; Ryoko, Nakai.; Yasuyuki, Kirii.; Koichi, Yokota.; Toshiji, Tada. J. Biochem. 2012, 151(5), 541-549.
CrossRef - Zhanzhan, Li.; Liangfang Shen, Na Li. Manag. Res. 2018, 10, 6905-6912.
CrossRef - Antonia, L. Pritchard.; Nicholas, K. Hayward. Clin .Cancer. Res. 2013,19(9), 2301-2309.
CrossRef - Eun, K. K.; Choi, E. Biochim. et .Biophys. Acta. 2010, 1802, 396- 405.
CrossRef - Parray, A. A.; Baba, R. A.; Bhat, H. F. Invest. 2014, 32(8), 416–422
CrossRef - Hua Liao, M. M.; Jia-li Kang, ; Wen-yan Jiang, M. M.; Cui Deng, M. M.; Jin Yuan, M. M.; Rong Shuai, M. M. Int. J. Gynecol. Cancer. 2015, 25, 1548-1556.
CrossRef - Vijesh Arun, A.M.; Isloor, M.; Sandeep Telkar T.; Arunmoli, Hoong-Kun Fun. Arab. J. Chem. 2013, 6, 197 – 204.
CrossRef - Bhargavi, M.; Sreekanth, S.; Sarita Rajender, P. Biol. Chem. 2017, 68, 43-55.
CrossRef - Nath, M.; Jairath, R.; Eng, G.; Song, X.; Kumar, J. Organomet. Chem. 2005, 690, 134-144.
CrossRef - Lokesh, M. R.; Krishnamurthy, G.; Bhojyanaik, H. S.; Shashikumar, N. D.; Murali, P. Krishna.; Sreekanth, B. Der. Pharma. Chemica. 2014, 6(6), 192 – 202.
- Pulin, Nath.; Shreedhar, D. D. Pharma. Chemica. 2014, 6(1), 253-261.
- Saritha, A.; Venkata Ramana Reddy, Ch.; Sireesha, B. J. Chemistry and Chemical Sciences. 2017, 7(2), 138-150.
- Shakir, M.; Mohammed, A. K.; Nasman, O. S. M. Polyhedron. 1996, 15, 3487-3492.
CrossRef - Agarwal, R. K.; Prasad, S. Chem. Appl. 2005, 3, 271-288.
CrossRef - Anupama, B.; Padmaja, M.; Gyana kumari, C. E- J. Chem. 2012, 9(1), 389-400.
CrossRef
- Djedouani, A.; Bendaas, ; Boufas, S.; Allain, M.; Bouet, G.; Khan, M. Acta. Cryst. 2007, E63, 1271-1273.

This work is licensed under a Creative Commons Attribution 4.0 International License.









