Synthesis, Characterization, Antimicrobial Activity and Molecular Docking Studies of New Benzimidazole, Benzoxazole, Imidazole and Tetrazole Derivatives
Department of Chemistry, SRM Institute of Science and Technology, Ramapuram Campus, Chennai-600089, Tamil Nadu, India.
Corresponding Author E-mail : helenkavithap2020@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/360411
Article Received on : 20 July 2020
Article Accepted on : 21 Aug 2020
Article Published : 06 Aug 2020
In the present research work, 12 new compounds, such as benzimidazole (3, 3a-c), benzoxazole(4), imidazole(5, 5a-c) and tetrazole(6, 6a-c) were synthesized. FT-IR, Proton NMR (1H), 13C-NMR, Mass spectral values were used to prove the structures of the compounds. The antimicrobial potential of the representative compounds was assessed using the disc diffusion process. All the benzoxazole, benzimidazole, imidazole and tetrazole derivatives prepared in this investigation show good antimicrobial activity. However the antimicrobial activities of the compounds are less compared with standard drugs. Molecular docking studies have also done for the synthesized compounds all the compounds show hydrogen bond interactions with receptor protein.
KEYWORDS:Antimicrobial Activity; Benzoxazole; Benzimidazole; Imidazole; Molecular Docking; Tetrazole
Download this article as:| Copy the following to cite this article: Arulmurugan S, Kavitha Helen P. Synthesis, Characterization, Antimicrobial Activity and Molecular Docking Studies of New Benzimidazole, Benzoxazole, Imidazole and Tetrazole Derivatives . Orient J Chem 2020;36(4). |
| Copy the following to cite this URL: Arulmurugan S, Kavitha Helen P. Synthesis, Characterization, Antimicrobial Activity and Molecular Docking Studies of New Benzimidazole, Benzoxazole, Imidazole and Tetrazole Derivatives . Orient J Chem 2020;36(4). Available from: https://bit.ly/3kh6sRE |
Introduction
The present situation is that the incidence of multidrug-resistant bacterial infections and physicians has become dependent on vancomycin is an antibiotic used to treat a number of complicated infections that are immune to conventional agents, suggesting that new groups of antimicrobials need to be created.1 Subsequently, antimicrobial agents whose chemical properties vary fully from those current agents need to be built up and can replace the resistance problem.2
Benzimidazole based compounds are of broad area of interest as a result of their various biological activities like antimicrobial activity,3 antioxidant activity,4 anticancer activity,5 antidiabetic activity,6 antiviral activity,7 anticonvulsant activity8 etc.
Benzoxazole is one of the main heterocyclic compounds that are very useful in the area of medicine. Among other medicinal compounds it has been used, making it a convertible heterocyclic compound with an extensive variety of biological activities, such as antimicrobial activity,9 antiinflammatory activity,10 anticancer activity,11 aminopeptidase activity,12 anti-HIV13 etc.
Imidazole is standard five-membered heterocyclic scaffolds containing nitrogen, and is commonly found in natural products and medicinal molecules. In addition, heterocyclic imidazole-based compounds possessing various biological activities, such as antibacterials14, antifungal,15 anti-inflammatory,16 antiviral,17 anti-parasitic,18 anticancer,19 antihistaminic,20 and enzyme inhibition.21
In recent decades, the synthesis of tetrazoles and the study of their chemical and biological behavior have become more important in the biological and pharmaceutical fields such as antibacterial,22 antifungal,23 anticancer,24 anti-inflammatory25 and analgesic26 activities. In the above-mentioned facts it was anticipated that, when mixed together, these active pharmacophores would produce novel molecular compounds that are likely to exhibit fascinating biological properties. Subsequently, continuation of our attention to the synthesis of biologically active heterocycles,27 we have reported some new heterocyclic compounds like benzimidazole, benzoxazole, imidazole and tetrazole derivatives synthesis and antimicrobial evaluation.
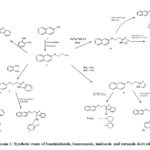 |
Scheme 1: Synthetic route of benzimidazole, benzoxazole, imidazole and tetrazole derivatives. |
Experimental Techniques
Infrared spectra were taken on the Perkin-Elmer FT-IR 1600 spectrometer using potassium bromide disks. NMR spectra were recorded by Bruker spectrometer at 500 MHz and 125 MHz for 13C spectra. Melting points have analyzed by a digital melting point apparatus. The reactions were monitored by TLC using solvent systems of different polarities. Mass spectra recorded by JEOL GCmate spectrometer.
Synthesis of Heterocyclic Compounds
Synthesis of heterocyclic compounds, such as 3, 4, 5, 6 were prepared according to the reported procedure27. Synthesis of 3a, 3b, 5a, 5b, 6a, 6b were synthesis using reported procedure28 and the synthesis of 3c, 5c, 6c derivatives were prepared according to reported procedure.29
 |
Table 1: Melting Point and Yield of the synthesized compounds |
Biological Activity Studies
Determination of Antimicrobial Study
Antimicrobial properties of the samples were screened against bacteria’s, such as S. aureus, P. aeruginosa, and B. subtilis and fungi, such as C. albicans and A. niger by Disc Diffusion Method. Muller Hinton agar media and PDA (Potato Dextrose Agar) used antibacterial and antifungal studies to subculture different strains of microorganisms, respectively.
Molecular Docking Tools
Docking studies were performed for synthesized compounds according to the reported procedure 30with target proteins by Glide 5.5 module of Schrodinger suite.
Result and Discussion
Table 2: Spectral data of synthesized compounds
|
Compound No |
FT-IR (KBr) cm-1 |
1H NMR(DMSO-d6) ( d ppm) |
13C NMR(MeOD) |
Mass spectra (m/z %) |
|
3 |
3055(Ar-CH), 2942(CH2), 1628(C=N), 3403(-NH). |
7.8 (m, 4H, Aromatic-H, (Benzimidazole ring)), 7.35 (m, 3H, Aromatic-H4,5,8), 7.19 (dd, 1H, Aromatic-H6), 7.12 (m, 1H, Aromatic-H7), 7.08 (dd, J=11 Hz, 1H, Aromatic proton-H3), 5.0 (s, 1H, NH group Benzimdazole ring), 7.74 (m, 1H, Aromatic-H1), 3.07 ( t, 2H, Methylene proton, -O-CH2-CH2 group), 4.30 (t, 2H, Methylene proton, -O-CH2–CH2 group) |
– |
288.13 [M+1] |
|
3a |
1628(-C=N), 1363(-C-N), 1120(-C-O-C-), 2941(CH2), 3056(Ar-CH). |
7.76 (m, 4H, Aromatic-H, (Benzimidazole ring)), 7.33 (m, 3H, Aromatic-H4,5,8), 7.43 (m, 1H, Aromatic-H1), 7.17 (dd,1H, Aromatic-H6), 7.25 (d, 1H, Aromatic-H7), 2.98 (t, J=12 Hz, 2H, Methylene proton, -O-CH2-CH2 group), 4.32 (t, 2H, Methylene proton, -O-CH2-CH2 group), 4.8 (s, 2H, -CH2-N piperidine ring), 2.41 (t, 2H, -CH2-N-C- piperidine ring), 7.09 (dd,1H, Aromatic-H3), 1.71 (t, 2H, CH2-CH2-C- piperidine ring), 3.70 (t, 2H, -CH2-CH2-C- piperidine ring). |
155.9, 153.3, 134.6, 129.4, 129.2, 127.2, 126.5, 126.1, 123.6, 118.1, 117.8, 106.8, 67.3, 62.8, 55.5, 25.6, 24.0, 17.6. |
385.09[M+1] |
|
3b |
1628(-C=N), 1323(-C-N), 1116(-C-O-C-), 2942(CH2), 3055(Ar-CH). |
3.08 (t, J=10 Hz, 2H, Methylene proton,-CH2-CH2-C), 2.3 (s, 2H, Methylene proton, -CH2-N-CH2– morpholine ring), 4.8(s, 2H, Methylene proton,-N-CH2-N- morpholine ring), 4.31 (t, J=10 Hz, 2H, Methylene proton, -CH2-CH2-), 3.38 (s, 2H, Methylene proton,-CH2-O-CH2-), 7.82 (m, 4H, Aromatic-H, (Benzimidazole ring)), 7.74 (m, 1H, Aromatic-H1), 7.39 (m, 3H, Aromatic-H4,5,8), 7.23 (dd,1H, Aromatic-H7), 7.50 (m, 1H, Aromatic-H6), 7.08 (dd,1H, Aromatic-H3) |
– |
386.62 [M+1] |
|
3c |
1627(-C=N), 1682(-C=O), 1323(-C-N), 1104(-C-O-C), 2919(CH2), 3027(Ar-CH), 772(C-Cl) |
7.79 (m,4H, Aromatic-H, (Benzimidazole ring)), 7.36 (m, 3H, Aromatic-H4,5,8), 7.19 (m, 1H, Aromatic-H6), 7.06 (m,1H, Aromatic-H3), 7.28 (m, 1H, Aromatic-H7), 4.32 (t, 2H, Methylene proton, –O-CH2-CH2-group), 7.56 (m, 1H, Aromatic-H1), 2.98 (t, 2H, Methylene proton, -CH2-CH2-CN group), 7.65 (m, 4H, Aromatic proton, Phenyl group). |
165.7, 155.9, 140.9, 137.2, 134.6, 129.6, 129.4, 129.2, 129.1, 128.9, 128.4, 127.8, 127.2, 127.2, 127.1, 126.5, 126.5, 126.1, 125.9, 125.3, 123.6, 123.3, 121.9, 118.4, 118.1, 117.8, 107.2, 106.8, 65.5, 62.8, 62.7, 28.9, 17.6. |
428.02 |
|
4 |
1628 (–C=N), 1120 (C-O-C), 2942 (-CH2), 3055 (Ar-CH) |
7.73 (m, 4H, Aromatic-H, (Benzoxazole ring)), 7.38 (m, 3H, Aromatic-H4,5,8), 7.11 (d, 1H, H6), 7.07 (dd, 1H, Aromatic-H7), 7.64 (d, 1H, H3), 4.30 (t, 2H, Methylene proton,-O-CH2-CH2-), 7.27 (m, 1H, Aromatic-H1), 2.85 (t, 2H, Methylene proton, -CH2-CH2) |
164.0, 154.9, 135.0, 128.9, 128.5, 127.2, 125.8, 125.7, 122.5, 117.8, 108.5, 64.0, 27.2 |
289.1 [M+1] |
|
5 |
3320 (-NH), 3056 (Ar-CH), 2942 (CH2), 1628(C=N), 1120 (C-O-C). |
7.86 (m, 3H, Proton 4,5,8 Aromatic ring), 7.38 (s, 2H, Aromatic proton H6,7), 7.22 (d, 1H, Aromatic-H3), 7.49 (t, 1H, Aromatic-H1), 3.09(s, 2H, Methylene proton,CH2-CN), 4.32 (s, 2H, Methylene protons, -O-CH2), 9.8 (s, 1H, NH-Imidazole ring), 6.73 (d, 1H-Imidazole ring.), 6.66 (d,1H-Imidazole ring) |
– |
238.1 [M+1] |
|
5a |
1628(C=N), 1338(C-N), 1120(C-O-C), 2942(CH2), 3056(Ar-CH) |
1.68 (m, 2H, -CH2-CH2–CH2, piperidine), 3.01 (t, 2H, Methylene protons, -CH2-N-CH2– piperidine), 7.26 (d, 1H, Aromatic-H3), 4.33 (t, 2H, -O-CH2-CH2-), 1.42 (m, 2H, Methylene protons, -CH2-CH2-CH2– piperidine), 7.76 (m, 3H, Aromatic-H4,5,8), 7.34 (m, 1H, Aromatic-H1), 7.43 (m, 1H, Aromatic-H6), 7.17 (dd, 1H, Aromatic-H7), 6.87 (d, 1H, proton Imidazole ring), 4.78 (s, 2H, Methylene protons, N-CH2-N), 7.01 (d, 1H, proton Imidazole ring). |
155.9, 155.3, 144.6, 134.6, 129.4, 129.2, 127.2, 126.5, 126.1, 123.6, 118.1, 117.8, 106.7, 70.1, 67.2, 62.8, 55.1, 26.1, 25.1, 24.0, 17.6 |
334.92 [M+1]. |
|
5b |
1628(C=N), 1338(C-N), 1119(C-O-C), 2942(CH2), 3056(Aromatic-CH) |
7.77 (m, 2H, Proton 4,5,8, Aromatic ring), 7.43 (m, 1H, Aromatic-H5), 7.34 (m, 2H, Aromatic-H6,7), 7.26 (d, 1H, Aromatic-H1), 7.17 (dd, 1H, Aromatic-H3), 4.30 (t, 2H, O-CH2-CH2), 2.99 (t, 2H, CH2-CH2-C=N), 6.81 (d,1H, -CH proton, Imidazole ring), 6.90 (d, 1H, -CH proton, Imidazole ring), 3.38 (t, 2H, Methylene protons , -CH2-O-CH2-), 2.46 ( t, 2H, Methylene protons, -CH2-N-CH2-), 4.72 (s, 2H, Methylene protons, N-CH2-N morpholine ring) |
155.9, 143.5, 134.6, 129.4, 129.2, 127.2, 126.6, 126.1, 123.6, 118.1, 117.8, 106.8, 69.0, 66.4, 62.8, 55.1, 17.6 |
337.18[M+1] |
|
5c |
1629(C=N), 1689(C=O), 1358(C-N), 1120(C-O-C), 2942(CH2), 3055(Ar-CH), 792(C-Cl) |
7.77 (m, 4H, Aromatic-H, Phenyl group), 7.5 (m, 3H, Aromatic-H4,5,8), 7.26 (d, 1H, Aromatic-H1), 7.17 (dd, 1H, Aromatic-H3), 7.34 (m, 2H, Aromatic-H6,7), 6.99 (d, 2H, Aromatic proton-Imidazole ring), 4.31 (t,2H, Methylene protons, O-CH2-C- group), 2.99(t, 2H, Methylene protons, -CH2-CH2-CN group) |
164.0, 155.9, 144.5, 134.6, 132.7, 132.1, 130.8, 130.5, 129.4, 129.2, 127.2, 126.5, 126.5, 126.1, 123.6, 118.1, 117.8, 106.8, 62.8, 17.6. |
378.49 [M+2] |
|
6 |
1442(N=N), 1160(N-N=N), 1600 (C=N), 1120 and 1043 (Tetrazole ring), 1358(C-O-C), 2941(CH2), 3056(Ar-CH), 3423(-NH) |
7.66 (m, 3H, Aromatic-H4,5,8), 7.38 (s, 2H, Aromatic-H6,7), 7.20 (d, 1H, Aromatic-H3), 7.46 (d, 1H, Aromatic-H1), 3.09 (s, 2H, Methylene protons, CH2-CH2-CN), 4.32 (s, 2H, Methylene protons, O-CH2-CH2), 9.78(s, 1H, NH proton-Tetrazole ring) |
– |
|
|
6a |
3052 (Ar-CH),1113(C-O-C), 2852(CH2), 1621(C=N), 1415 |
7.66 (d, 1H, Aromatic-H1), 7.03 (d, 1H, Aromatic-H3), 7.73 (d, 2H, Aromatic-H4,5,8) 7.86 (d, Aromatic-H5), 7.25 (t, 1H, Aromatic-H6), 7.42 (m,1H, Aromatic-H7) 4.16 (s,2H, -O-CH2-CH2-), 1.67 (m, 2H, Methylene protons, -CH2-CH2-CH2-), 1.29 (s,2H, Methylene protons, CH2-CH2-C-N), 2.66 (s, 2H, Methylene protons, CH2-N-CH2), 1.57 (s,2H, Methylene protons, CH2-CH2-CH2 piperidine), 6.12 (s,2H, Methylene protons, N-CH2-N) |
159.3, 156.4, 132.9, 128.7, 128.5, 128.3, 125.9, 122.0, 120.8, 118.5, 110.9, 62.9, 56.5, 53.5, 25.5, 23.5, 17.1 |
337.60 [M+1] |
|
6b |
3050 (Ar-CH), 1115(C-O-C), 2922(CH2), 1622(C=N), 1416 (-N=N), 1232 (-N-N=N), 1031 and 1076 (Tetrazole ring) |
7.68 (d, 1H, Aromatic-H1), 7.04 (d, 1H, Aromatic-H3), 7.74 (d, 2H, Aromatic-H4,5,8), 7.93 (d, 1H, Aromatic-H5), 7.26 (t,1H, Aromatic-H6), 2.66 (s, 2H, Methylene protons, CH2-N-CH2), 7.42 (m,1H, Aromatic-H7), 3.33 (s, 2H, Methylene protons, CH2-CH2-C=N), 4.14 (s, 2H, Methylene protons, O-CH2-CH2), 3.73 (t, 2H, Methylene protons, CH2-O-CH2), 6.1 (s, 2H, Methylene protons, N-CH2-N). |
159.5, 155.4, 133.1, 128.9, 128.8, 128.3, 126.1, 122.3, 121.3, 118.0, 111.2, 67.1, 66.8, 66.5, 55.1, 52.8, 25.0. |
339.33 [M+1] |
|
6c |
3054 (Ar-CH), 1119(-C-O-C), 2920(Aliphatic-CH2), 1628(C=N), 1466 (-N=N), 1258 (-N-N=N), 1119 and 1045 (Tetrazole ring) |
3.00 (t, 2H, Methylene protons, -CH2-CH2-CN), 4.32 (t, 2H, Methylene protons, O-CH2-CH2), 7.13(dd, 1H, Aromatic-H1), 7.18 (dd, 1H, Aromatic-H3), 7.24 (m, 3H, Aromatic-H, phenyl), 7.34 (m, 1H, Aromatic-H4,5,8), 7.43 (m, 1H, Aromatic-H6,7), 7.30 (m, 2H, Aromatic-H5) |
174.1, 161.6, 156.7, 155.9, 140.9, 134.8, 134.6, 130.7, 130.0, 129.6, 129.4, 129.2, 129.1, 129.1, 128.9, 127.2, 127.1, 126.6, 126.5, 126.1, 125.8, 123.6, 123.2, 118.5, 118.1, 112.4, 106.8, 106.4, 66.3, 62.8, 54.3, 25.3, 17.6. |
379.12[M+2] |
Antimicrobial Activity
The antimicrobial evaluation of the representative samples was performed by Disc Diffusion Technique. The test bacteria’s, such as S. aureus, B. subtilis, P. valgaris and K. aerogenes as well as fungi, such as A. niger and C. albicans. Test effect distinguished with positive control (Ciprofloxacin for bacteria; Nystatin for fungi). The values of zone of inhibitions are presented in the table (Table 3). The photographs showing the zone of inhibition against the tested species are given in the figures (Fig. 1)
Table 3: Antimicrobial study of the new compounds
|
S. No |
Microorganism |
Zone of Inhibition (mm) |
|||||||
|
3a |
3b |
5b |
5c |
6a |
6b |
Solvent control |
Std |
||
|
1 |
Staphyloccocus aureus |
18 |
20 |
15 |
10 |
14 |
19 |
– |
35 |
|
2 |
Bacillus subtilis |
16 |
20 |
14 |
10 |
12 |
20 |
– |
40 |
|
3 |
Proteus valgaris |
15 |
12 |
16 |
13 |
14 |
12 |
– |
26 |
|
4 |
Klebseilla aerogenes |
16 |
16 |
20 |
12 |
16 |
18 |
– |
30 |
|
5 |
Aspergillus niger |
18 |
20 |
20 |
18 |
16 |
18 |
– |
35 |
|
6 |
Candida albicans |
21 |
20 |
24 |
20 |
20 |
21 |
– |
32 |
It is concluded from the table ( Table 3) that all the compounds used for the antimicrobial analysis displayed moderate to strong activity against both the bacteria and fungi strains compared to the standard drug. Table 2 compounds have again been tested for Minimum Inhibitory Concentration taking different concentrations, such as 500μg / ml, 250μg / ml, and 125μg / ml. It is found from Table 3, that all compounds exhibited inhibition at 125μg / ml. Figures display the images showing the Minimum Inhibitory Concentration (Fig 2).
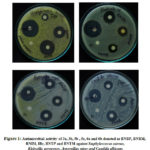 |
Figure 1: Antimicrobial activity of 3a, 3b, 5b , 5c, 6a and 6b denoted as BNBP, BNBM,BNIM, |
Table 4: Minimum inhibitory concentration of the new compounds
|
S. No |
Microorganism |
Zone of Inhibition(mm) |
|||||
|
Sample code |
125µg/ml |
250µg/ml |
500µg/ml |
Solvent control |
Std |
||
|
1 |
Staphyloccocus aureus |
3b |
14 |
16 |
22 |
– |
35 |
|
2 |
Bacillus Subtilis |
3b |
15 |
18 |
20 |
– |
40 |
|
6b |
12 |
14 |
20 |
– |
30 |
||
|
3 |
Klebseilla aerogenes |
5b |
17 |
18 |
21 |
– |
30 |
|
4 |
Aspergillus niger |
3b |
16 |
18 |
22 |
– |
35 |
|
5b |
19 |
20 |
21 |
– |
|||
|
5 |
Candida albicans |
3a |
12 |
16 |
20 |
– |
32 |
|
3b |
18 |
18 |
21 |
– |
|||
|
5b |
16 |
16 |
20 |
– |
|||
|
5c |
18 |
19 |
20 |
– |
|||
|
6a |
18 |
20 |
24 |
– |
|||
|
6b |
16 |
20 |
20 |
– |
|||
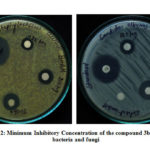 |
Figure 2: Minimum Inhibitory Concentration of the compound 3b against bacteria and fungi |
Molecular Docking Study
Docking experiments were conducted using Schrodinger (Mastro 9.2 V) suite to learn the right binding site of the compounds. All the compounds from Scheme-1, which were tested for the antimicrobial activity were taken for the docking studies and molecular docking was performed against antimicrobial protein beta-ketoacyl-acp synthase III+ malonyl-COA (pdb id: 1HNJ). All the ligands interact with antimicrobial protein receptor. Glide Score, glide energy and hydrogen bond distance for the Compounds with protein are given in the Table.4.
Amino acid residue ARG36-NH of the enzyme beta-ketoacyl-acp synthase III is form hydrogen bond with the O atom of the ligand 3b (Fig 3). Fig. 3 shows that the amino acid residues ARG249, PHE213 and TRP32 of the enzyme beta-ketoacyl-acp synthase III are involved in π-π interaction with benzene of the benzimidazole ring and chlorobenzene ring of the ligand 3c, and the ARG36-NH of the amino acid residue form hydrogen bond with compound 3c.
Amino acid residue ASN247-NH of the enzyme beta-ketoacyl-acp synthase III is formation of hydrogen bond interaction with the O atom of 2-Napthol ring of the compound 4. The N atom of the Imidazole compound 5a is involved in hydrogen bond interaction with O atom of ASN247. Similarly the Nitrogen atom of Imidazole compound 5b is produced hydrogen bond interaction with O atom of ASN247. The N of tetrazole compound 6a is hydrogen bond with O atom of ASN247 amino acid residue. The amino acid residue ARG249-NH of the enzyme beta-ketoacyl-acp synthase is formed hydrogen bond interaction with O atom of the morpholine ring of the compound 6b. The amino acid residues ARG49-O, PHE18-O, ARG36-N and TRP32-O of the enzyme beta-ketoacyl-acp synthase are involved in hydrogen bond interaction with N,N, C=O, benzene (π-π) of ligand compound 6c.
Table 5: Glide Score, Glide Energy and Hydrogen Bond Distance for the Compounds with Protein beta-ketoacyl-acp synthase III+ malonyl-COA (pdb id: 1HNJ)
|
Comp. code |
Glide gscore |
Glide energy kcal/mole |
Hydrogen bond |
|
|
Donor |
Acceptor |
|||
|
3b |
-6.44 |
-38.71 |
NH of ARG36 |
O of (Morpholine ring) |
|
3c |
-6.14 |
-44.89 |
O of carbonyl group |
NH of ARG36 |
|
Benzene ring |
ARG249(π-π) |
|||
|
PHE213(π-π) |
||||
|
TRP32(π-π) |
||||
|
4 |
-6.79 |
-37.40 |
NH of ASN247 |
O of 2-Napthol |
|
5a |
-6.59 |
-37.38 |
N (Imidazole ring) |
O of ASN247 |
|
5b |
-6.33 |
-39.01 |
N of Imidazole |
O of ASN247 |
|
6a |
-6.65 |
-39.49 |
N (tetrazole ring) |
O of ASN247 |
|
6b |
-6.20 |
-31.25 |
NH ARG 249 |
O of (Morpholine ring) |
|
6c |
-6.47 |
-37.20 |
N of Terazole ring |
O of ARG 49 |
|
N of Terazole ring |
O of PHE18 |
|||
|
NH of ARG36 |
C=O of tetrazole derivatives |
|||
|
O of TRP-32 |
CH of benzene ring |
|||
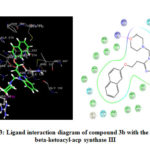 |
Figure 3: Ligand interaction diagram of compound 3b with the enzyme beta-ketoacyl-acp synthase III |
Conclusion
In the current research work, 12 new heterocyclic compounds, such as benzimidazole, benzoxazole, imidazole and tetrazole derivatives were synthesized from 2-Napthol. In the expected region, the FT-IR spectra of newly synthesized derivatives showed absorption bands. The 1H-NMR and 13C-NMR spectra provided believable proof for the structure assigned to the synthesized compounds. Complete proof was obtained for the structures by getting the mass spectrum that reveal molecular ion peak equivalent to the molecular weight. Some of the representative compounds have been tested for antibacterial study against bacteria, such as S. aureus, B. subtilis, P. vulgaris and K. aerogenes by disc diffusion method. The antifungal property of the compounds was tested against C. albicans and A. niger. The outcome of the antimicrobial study indicates that the tested compounds showed reasonable to good antimicrobial property. However, the activity of the compounds is less than the standard drug. The Molecular Docking study was carried out using Schrodinger (Maestro 9.2v) software. The compounds were screened for the in vitro antibacterial study was screened in silico study against the antimicrobial protein enzyme beta-ketoacyl-acp synthase III. The findings of the analysis suggest that the compounds we used for docking exhibited interactions between the hydrogen bonds.
Acknowledgment
The authors are very much grateful for the moral support and encouragement offered by the management of the SRM Institute of Science and Technology, Ramapuram Campus.
Conflict of Interest
No conflict of interest.
References
- Chilumula, N. R.; Gudipati, R.; Ampati, S.; Manda, S.; Gadhe, D. J .Chem. Res. 2010, 1, 1-6
- Dawood, N. T. A Chem. Pharm. Res, 2011, 3, 111-121
- Kumar, B. V.; Vaidya, S. D.; Kumar, R. V.; Bhirud, B.; Mane, R. B. Eur J Med Chem. 2006, 41, 599-604
- Kus, C.; Kilcigil, G. A.; Eke, B .C. Arch Pharm Res. 2004, 27, 156-163
- Kawasaki, K.; Masubachi, M.; Morikami, K.; Sogebe, S.; Aoyama, T.; Ebiike, H.; Niizuma, S.; Hayase, M.; Fujii, T.; Sakata, K.; Shindoh, H.; Shiratori, Y.; Aoki, Y.; Ohtsuka, T.; Shimma, N. Med. Chem. Lett. 2003, 13, 87-91
- Bathini, P.; Kameshwari, L.; Vijaya, N. J. Basic. Clin. Pharmacol. 2013, 2, 814-818
- Fonseca, T.; Gigante, B.; Marques, M.M.; Gilchrist, T.L.; Clercq, E.D. Med. Chem. 2004, 12, 103-112
- Falco, J.L.; Pique, M.; Gonzalez, M.; Buira, I.; Mendez, E.; Terencio, J.; Perez, C.; Princep, M.; Palomer, A.; Guglietta, A. J. Med. Chem. 2006, 41, 985-990
- Arpaci, O.T.; Oren, I.; Altanlar, N. II Farmaco. 2002, 57, 175-181
- Ampati, S.; Jukanti, R.; Sagar, V.; Ganta, R.; Manda, S. Der Chemica Sinica. 2010, 1, 157-168
- McKee, M. L.; Kerwin, S. M. Med. Chem. 2008, 16, 1775-1783
- Cellier, M.; Fabrega, O.J.; Fazackerley, E.; James, A.L.; Orenga, S.; Perry, J.D.; Salwatura, V. L.; Stanforth, S. P. Med. Chem., 2011, 19, 2903-2910
- Medebielle, M.; Ait-Mohand, S.; Burkhloder, C.; Dolbier Jr, W. R.; Laumond, G.; Aubertin, A. Fluor. Chem. 2005, 126, 533-540
- Khabnadideh, S.; Rezaci, Z.; Khalafi, N.A.; Motazedian, M.H.; Eskandari, M. DARU, 2007, 15, 17-20
- Rezaei, Z.; Khabnadideh,S.; Zomorodian, K.; Pakshir, K.; Kashi, G.; Sanagoei, N.; Gholami, S. Pharm. Chem. Life Sci. 2011, 344, 658-665
- Che, H.; Tuyen, T.N.; Kim, H.P.; Park, H. Med. Chem. Lett. 2010, 20, 4035-4037
- Ren, J.S.; Nichols, C.; Bird, L.E.; Fujiwara, T.; Sugimoto, H.; Stuart, D.I. Biol. Chem. 2000, 275, 14316-14320
- Kapoor, V.K.; Chadha, R.; Venisetty, P.K.; Prasanth, S. Sci. Ind. Res. 2003, 62, 659-665
- Zhang, M.; Ding, Y.; Qin, K. X.; Xu, Z.G.;Lan, H. T.; Yang, D. L.; Yi, C. Diver. 2019, 1-8
- Kitbunnadaj, R.; Zuiderveld, O.P.; Christophe, B.; Hulscher, S.; Menge,W.M.; Gelens, E.; Snip, E.; Bakker, R.A.; Celanire, S.; Gillard, M. Med. Chem., 2004, 47, 2414-2417.
- Hille, U.E.; Zimmer, C.; Vock, C.A.; Hartmann, R.W. ACS Med. Chem. Lett. 2011, 2, 2-6
- Zhang, H.Z.; Gan, L.L.; Wang, H.; Zhou, C.H. Mini Rev. Med. Chem., 2017, 7, 122-166
- Nami, S.; Aghebati-Maleki, A.; Morovati, H.; Aghebati-Maleki, L. Pharmacother, 2019, 110, 857-868
- Popova, E.A.; Protas, A.V.; Trifonov, R.E. Anti-Cancer. Med. Chem. 2017, 17, 1856-1868
- Maria Dorathi Anu, M.; Jayanthi, M.; Damodar Kumar, S.; Raja, S.; Thirunavukkarasu, S.V. J. ChemTech Res. 2013, 5, 1982-1990
- Rajasekaran, A.; Thampi, P.P. J. Med. Chem., 2004, 39, 273-279.
- Arulmuruggan, S.; Kavitha, H.P. Pharm. 2013, 63, 253-264.
- Praveena, C.H.L.; Rani, V.S.; Spoorthy, Y.N.; Ravindranath, L.K. Chem. Pharm. Res., 2013, 5, 280-292
- Rajasekaran, A.; Thampi, P.P. Pharm. Tur., 2003, 45, 235-240
- Ibrahim, F.; Nassar, Saad, R.; Atta-Allah, Abdel-Sattar, S.; Elgazwy, H. J. Enzyme. Inhib. Med. Chem. 2015, 30, 396–405

This work is licensed under a Creative Commons Attribution 4.0 International License.










