Synthesis, Characterization and Single Crystal Structure of Carbanionic Sigma Complexes of Sodium and Potassium Salts
M. Bhavya* and R. Malarvizhi
Department of Chemistry, Seethalakshmi Ramaswami College, Affiliated to Bharathidasan University, Tiruchirappalli-620002, Tamil Nadu, India
Corresponding Author E-mail : malarsri.kiruba@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/360402
Article Received on : 22 May 2020
Article Accepted on : 23-July-2020
Article Published : 28 Aug 2020
Barbituric acid derivatives are very much effective in anxiolytics, hypnotics, and anticonvulsant agents. In this context, in the present investigation, new molecular salts have been synthesized in the crystalline form. They have been characterized through UV-VIS, IR, 1H NMR, 13C NMR, and X- ray diffraction studies. In this anion, 1, 3-dimethylbarbituric acid ring and substituted dinitrophenyl ring, linked via a C-C bond.
KEYWORDS:FT-IR; NMR; Potassium Methoxide; Sodium Methoxide; UV, X-ray Diffraction
Download this article as:| Copy the following to cite this article: Bhavya M, Malarvizhi R. Synthesis, Characterization and Single Crystal Structure of Carbanionic Sigma Complexes of Sodium and Potassium Salts. Orient J Chem 2020;36(4). |
| Copy the following to cite this URL: Bhavya M, Malarvizhi R. Synthesis, Characterization and Single Crystal Structure of Carbanionic Sigma Complexes of Sodium and Potassium Salts. Orient J Chem 2020;36(4). Available from: https://bit.ly/2EKRqU4 |
Introduction
Synthetic nitroaromatic compounds widely used in a variety of materials, including drugs,1-3 pharmaceutical dyes,4-5 explosives,6-7 plastics, and pesticides. Several high energy density molecules are derived from nitroaromatic compounds.8-10 Nitroaromatic compounds are electron-deficient and react readily with electron-rich species. With electron-rich organic and inorganic bases, they undergo many types of reactions.11-14 Depends on the nature of electron-deficient nitroaromatic compounds and electron-rich species, they had three different types of interactions which occur, leading to the formation of Charge transfer complexes.15 Radical anion16 and Anionic sigma complexes.17
DNCB
(1-chloro-2, 4-dinitrobenzene) is a unique functional group used in medicinal chemistry. Nitro group is associated with mutagenicity and genotoxicity and therefore is often used in the drug discovery process.18 The nitroaromatic compound is also used to synthesize dyes and high energy materials. These compounds are very common environmental pollutants that are used as pesticides and explosives.19 DNCB molecule used in the preparation of many diverse organic molecules and biologically active molecules in recent years.
Barbituric Acid
(1,3-barbituric acid) has an active methylene group, i.e., (a cyclic molecule). It is expected to form a carbanion in the presence of a base. The C-5hydrogen atom substitution gives the complexes. Dyes involving active methylene group of barbituric acid have been considered in recent years.20, 21, 22, 23
Mechanism
Strong interaction between the base and the aromatic compound results in sigma adduct formation.24 A particular transfer of electronic charge from the base to the aromatic nucleus depleted of π electron density gives rise to the π-complex known as donor-acceptor or charge-transfer complex.25 The complete transfer of an electron from the base to the nitroaromatic compound results in the formation of radical anion.26 If the nitroaromatic bears a substituted alkyl group, carbanion is formed due to proton abstraction.27 The mechanism is given in scheme-I.
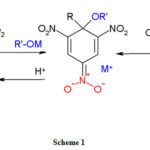 |
Scheme 1: |
Most of these reactions have been made many reviews written.28-34 Several reactions can proceed through cyclohexadiene ion of finite stability.35-37 As aromaticity is disrupted during the formation of the sigma complex, considerable changes occur in electronic configuration and hence visible spectroscopy has been a primary tool in deducing complex-forming reactions.38-40 The following mechanism is shown in Scheme- II.
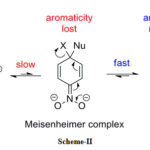 |
Scheme 2: Misenheimer complex |
The carbanionic sigma complex can also be made as sensors in many fields such as industry, environment, and biotechnology.
Experimental Methods
Synthesis of Molecular Salts
1-Chloro2,4-dinitrobenzene (DNCB) is dissolved in 30ml of ethanol be mixed with 1,3-dimethyl barbituric acid in 30ml of ethanol. Then added Sodium/potassium methoxide in 30ml of ethanol and shaken for 2-3 hrs. The excess ethanol be removed by distillation also the remaining solution was kept for one week when red colored crystals separated. These crystals recrystallized from distilled ethanol.
Thin Layer Chromatographic Studies
The study was carried out to check the purity of the isolated molecules. Silica gel was used as an adsorbent. The slurry of silica gel was made with chloroform and coated uniformly on a TLC plate. The complex solution was prepared in pure absolute ethanol and spotted on the plate. The eluent used was the mixture of tertiary butyl alcohol and ethyl acetate.
Results and Discussion of The Molecular Salts
In the present investigation, a red crystalline solid is obtained and the yield is 42% and 75%.
Crystal Structure Determination of Complexes
Crystal Structure of Sodium Salt
The amorphous solid of sodium salt is crystallized from ethylene glycol. The sodium cations are bridged through an oxygen atom of 1-chloro-2, 4-dinitro benzene, and 1, 3- dimethyl barbituric acid. In barbiturate residue bond lengths, bond angles are well-matched with that of barbiturate ion [42] and it shows the delocalization of negative charge. The crystal data represent selected bond angles, bond distances of sodium salt. According to X-ray crystallographic data, the isolated molecule is monoclinic system. The unit cell parameters having P121/n1 space group a=7.3269(5) Å, b=19.9241(13) Å, c=11.5431(9) Å, V=1665.1(2) Å3; Z=4.
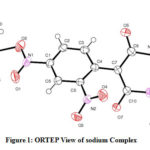 |
Figure 1: ORTEP View of sodium Complex |
Table 1: Bond angles (°)
|
O8-Na1-O9 |
163.11(17) |
O8-Na1-O10 |
83.38(15) |
|
O9-Na1-O10 |
107.24(18) |
O8-Na1-O10 |
105.47(16) |
|
O9-Na1-O10 |
89.58(17) |
O10-Na1-O10 |
79.92(13) |
|
O8-Na1-O2 |
92.73(14) |
O9-Na1-O2 |
82.65(17) |
|
O10-Na1-O2 |
156.10(15) |
O10-Na1-O2 |
78.44(12) |
|
O8-Na1-O3 |
90.31(13) |
O9-Na1-O3 |
75.29(15) |
|
O10-Na1-O3 |
99.05(12) |
O10-Na1-O3 |
163.88(16) |
|
O2-Na1-O3 |
104.56(12) |
O8-Na1-O1 |
70.05(13) |
|
O9-Na1-O1 |
95.85(16) |
O10-Na1-O1 |
149.52(13) |
|
O10-Na1-O1 |
120.71(12) |
O2-Na1-O1 |
44.35(9) |
|
O3-Na1-O1 |
67.40(10) |
O8-Na1-Na1 |
95.74(12) |
|
O9-Na1-Na1 |
100.81(14) |
O10-Na1-Na1 |
40.17(9) |
|
O10-Na1-Na1 |
39.75(8) |
O2-Na1-Na1 |
117.54(11) |
|
O3-Na1-Na1 |
137.02(11) |
O1-Na1-Na1 |
153.42(12) |
|
O8-Na1-H6O |
160.(3) |
O9-Na1-H6O |
22.(4) |
|
O10-Na1-H6O |
85.(4) |
O10-Na1-H6O |
88.(3) |
|
O2-Na1-H6O |
104.(3) |
O3-Na1-H6O |
76.(3) |
|
O1-Na1-H6O |
116.(4) |
Na1-Na1-H6O |
86.(4) |
|
N1-O1-Na1 |
87.9(2) |
N1-O2-Na1 |
104.5(3) |
|
N2-O3-Na1 |
127.3(3) |
Na1-O8-H2O |
117.(4) |
|
Na1-O8-H1O |
116.(5) |
H2O-O8-H1O |
89.(6) |
|
Na1-O9-H5O |
139.(7) |
Na1-O9-H6O |
73.(10) |
|
H5O-O9-H6O |
100.(10) |
Na1-O10-Na1 |
100.08(13) |
|
Na1-O10-H3O |
122.(6) |
Na1-O10-H3O |
104.(5) |
|
Na1-O10-H4O |
107.(5) |
Na1-O10-H4O |
115.(5) |
|
H3O-O10-H4O |
109.(7) |
Table 2: Bond Lengths
|
C8-O5 |
C8-O5 |
C8-O5 |
C8-O5 |
|
1.241(5) |
1.241(5) |
1.241(5) |
1.241(5) |
|
C8-N3 |
C8-N3 |
C8-N3 |
C8-N3 |
|
1.415(5) |
1.415(5) |
1.415(5) |
1.415(5) |
|
C9-N3 |
C9-N3 |
C9-N3 |
C9-N3 |
|
1.368(6) |
1.368(6) |
1.368(6) |
1.368(6) |
|
C9-N4 |
C9-N4 |
C9-N4 |
C9-N4 |
|
1.377(5) |
1.377(5) |
1.377(5) |
1.377(5) |
|
C10-O7 |
C10-O7 |
C10-O7 |
C10-O7 |
|
1.248(4) |
1.248(4) |
1.248(4) |
1.248(4) |
|
C10-N4 |
C10-N4 |
C10-N4 |
C10-N4 |
|
1.400(5) |
1.400(5) |
1.400(5) |
1.400(5) |
|
C11-H11A |
C11-H11A |
C11-H11A |
C11-H11A |
|
0.96 |
0.96 |
0.96 |
0.96 |
|
C11-H11B |
C11-H11B |
C11-H11B |
C11-H11B |
|
0.96 |
0.96 |
0.96 |
0.96 |
|
C11-H11C |
C11-H11C |
C11-H11C |
C11-H11C |
|
0.96 |
0.96 |
0.96 |
0.96 |
Crystal Structure of Potassium Salt
The structure of the potassium complex with DNCB, the crystals have been confirmed based on unit-cell contents. The crystal studies indicate that potassium cations are 8-coordinated and are bridged by an oxygen atom of 1,3-dimethyl barbituric acid. The K-O bond length range is 2.778-3.8699 (Shrivastava & Speakman, 1961). The Single-crystal XRD results have shown that the potassium salt of the present investigation possesses organometallic polymeric association. The crystal data represent selected bond distances, bond angles of potassium salt. The molecule is monoclinic crystal system having P21/n space group and cell parameters are a=11.3166(7) Å, b=7.2477 Å, c=20.3511(11) Å; Z=4.
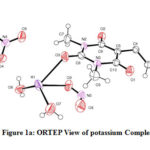 |
Figure 1a: ORTEP View of potassium Complex |
Table 1a. Bond angles (°).
|
C12-C1-C2 |
121.66(17) |
C12-C1-N5 |
118.92(1) |
|
O1-C10-N3 |
117.05(16) |
C5-C10-N3 |
116.22(16) |
|
C12-C11-C4 |
123.45(17) |
C12-C11-N4 |
114.88(16) |
|
C4-C11-N4 |
121.54(16) |
C1-C12-C11 |
118.11(17) |
|
O7-K1-O3 |
94.57(7) |
O7-K1-O6 |
137.71(7) |
|
O3-K1-O6 |
124.79(5) |
O7-K1-O6 |
73.77(6) |
|
O3-K1-O6 |
125.02(6) |
O6-K1-O6 |
92.26(6) |
|
O7-K1-O4 |
74.48(7) |
O3-K1-O4 |
159.63(5) |
|
O6-K1-O4 |
63.28(5) |
O6-K1-O4 |
69.12(6) |
|
O7-K1-O9 |
109.66(6) |
O3-K1-O9 |
71.94(5) |
|
O6-K1-O9 |
73.87(5) |
O6-K1-O9 |
162.89(6) |
|
O4-K1-O9 |
95.25(5) |
O7-K1-O4 |
129.72(6) |
|
O3-K1-O4 |
97.47(5) |
O6-K1-O4 |
65.55(5) |
|
O6-K1-O4 |
59.45(5) |
O4-K1-O4 |
102.74(4) |
|
O9-K1-O4 |
120.50(5) |
O7-K1-O8 |
70.96(6) |
|
O3-K1-O8 |
78.03(5) |
O6-K1-O8 |
100.18(6) |
|
O6-K1-O8 |
139.20(5) |
O4-K1-O8 |
82.13(5) |
|
O9-K1-O8 |
38.83(5) |
O4-K1-O8 |
159.31(5) |
|
O7-K1-K1 |
109.17(6) |
O3-K1-K1 |
145.66(4) |
|
O6-K1-K1 |
46.42(4) |
O6-K1-K1 |
45.83(4) |
|
O4-K1-K1 |
54.47(4) |
O9-K1-K1 |
119.57(5) |
|
O4-K1-K1 |
48.27(3) |
O8-K1-K1 |
132.51(5) |
|
O7-K1-H2O |
12.6(7) |
O3-K1-H2O |
105.8(7) |
|
O6-K1-H2O |
128.5(7) |
O6-K1-H2O |
62.8(7) |
|
O4-K1-H2O |
65.7(7) |
O9-K1-H2O |
118.2(7) |
|
O4-K1-H2O |
121.0(7) |
O8-K1-H2O |
79.5(7) |
|
K1-K1-H2O |
96.6(7) |
C8-N2-C6 |
123.46(16) |
|
C8-N2-C7 |
117.88(16) |
C6-N2-C7 |
118.55(16) |
|
C8-N3-C10 |
124.37(15) |
C8-N3-C9 |
117.53(16) |
|
C10-N3-C9 |
118.10(16) |
O5-N4-O4 |
123.19(18) |
|
O5-N4-C11 |
118.60(16) |
O4-N4-C11 |
118.15(17) |
|
C8-O3-K1 |
126.35(14) |
N4-O4-K1 |
145.39(15) |
|
N4-O4-K1 |
111.01(13) |
K1-O4-K1 |
77.26(4) |
|
K1-O6-K1 |
87.74(6) |
K1-O6-H4O |
107.(2) |
|
K1-O6-H3O |
105.(2) |
K1-O6-H3O |
134.(3) |
|
K1-O7-H1O |
118.(3) |
H4O-O6-H3O |
101.(3) |
|
H1O-O7-H2O |
131.(2) |
K1-O7-H2O |
109.(3) |
|
N5-O9-K1 |
102.(4) |
N5-O8-K1 |
90.75(13) |
|
O8-N5-C1 |
106.82(14) |
O8-N5-O9 |
122.32(18) |
|
119.45(19) |
O9-N5-C1 |
118.23(19) |
Table 2a: Bond Lengths
|
K1-O3 |
2.7228(16) |
K1-O6 |
2.778(2) |
|
K1-O6 |
2.8058(18) |
K1-O4 |
2.9608(19) |
|
K1-O9 |
3.0265(18) |
K1-O4 |
3.2287(17) |
|
K1-O8 |
3.3488(19) |
K1-K1 |
3.8699(10) |
UV-Visible Analysis
In the UV-Vis spectrum, the two peaks are formed with λmax values of 287.30nm, 484.34nm, and 245.93 nm, 446.86 nm (Fig 2,2a).
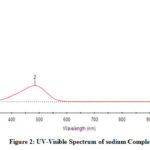 |
Figure 2: UV-Visible Spectrum of sodium Complex |
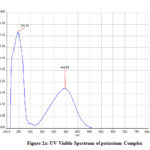 |
Figure 2a: UV-Visible Spectrum of potassium Complex |
FT-IR Analysis
FT-IR spectra are shown in (Fig 3,3a). NO2 asymmetric and symmetric stretching frequency is 1536 cm-1, 1532 cm-1 and 1331 cm-1 ,1327 cm-1. Here, C-Cl band is absent in the synthesized molecule The broad band observed between ~3600-2100 cm-1 is characteristic of amine salt.43 Carbonyl stretching frequency of 1,3-diethyl barbituric acid 1680 cm-1 and 1670 cm-1 during the formation of the complex.
 |
Figure 3: FT-IR Spectrum of sodium Complex |
 |
Figure 3a: FT-IR Spectrum of Potassium Complex |
NMR Analysis
1H NMR spectra are depicted in (Fig 4, 4a). This shows two peaks characteristics of the ring protons, one at δ 8.1ppm (S, 2H) and another at δ 8.3 ppm (S, 1H) are observed. The peak corresponding to six methyl protons appears at δ 3.1 ppm. 13C NMR spectra of molecular salts are presented in (Fig 5, 5a). 13C NMR spectrum indicate ten different carbon environments in the molecule. The peaks at 141.5 ppm and 87.1 ppm represents formation of C=C.
 |
Figure 4: 1H-NMR Spectrum of Sodium Complex |
 |
Figure 4a: 1H-NMR Spectrum of Potassium Complex |
 |
Figure 5: 13C NMR Spectrum of Sodium Complex |
 |
Figure 5a: 13C NMR Spectrum of potassium complex |
Conclusion
The present study focused on the characterization through various studies of carbanionic sigma complexes. Spectral data are inconsistent with the putative structure and further evidenced by single crystal XRD data. Various types of interactions possible between electron deficient compounds nitro aromatic compounds and bases are described. The cation and anion moiety are connected by hydrogen bonds. The complex for this work is stable in nanosize. Therefore, these may be changed into a potent drug in future.
Acknowledgement
I express my wholehearted gratitude to my eminent Research Supervisor Dr.R. Malarvizhi, Department of Chemistry, for scientific advice, knowledge sharing and many insightful discussions and suggestions. I am grateful thanks to the Secretary, Mr. R. Panchapakesan, and Dr.R. Padmavathy, Principal, Seethalakshmi Ramaswami College,Tiruchirappalli – 620 002, for having given me an opportunity to do research with necessary infrastructure and resources to carry out my work. I extend my gratitude to Dr.R.Vasuki, Head, Department of Chemistry, Vice Principal and SF-In Charge of Seethalakshmi Ramaswami College, Tiruchirappalli – 620 002, for constant encouragement and moral support.
Conflict of Interest
The authors declare no conflict of interest.
References
- Tripathi, K.D., Essentials of Medical Pharmacology, 6th, Jaypee Brothers Medical Publishers, Chennai 2009
- Ravikumar, K., Sridhar, B., Nanubolu, J.B., Hariharakrishnan, V., Singh, A.N. Acta Cryst, C68, O302
CrossRef - Elis-davies, G.C.R. Bellistein Org.Chem., 2013 , 9, 64
CrossRef - Shenai, V.A. Technology of Dyeing, Sevak publications, Vol-I, 1984
- Chatwal, G.R. Synthetic Dyes.,2nd, Himalaya Publishing Shouse, NewDelhi 1990
- Akhavan, J. The Chemistry of Explosives Royal Society of Chemistry: Great Britain, 2nd, K, 2004
- Agarwal, J.P., Hodgson, R.D. Organic Chemistry of Explosives, Willey, 2007
CrossRef - Urbanski, T. Chemistry of technology of Explosives, Pergamon Press, Oxford, Vol.1, 467, 1964
- Akhavan, J. The Chemistry of Explosives, 2nd, Royal Society of Chemistry: Cambridge, UK,2004
- Agarwal, J.P., Hodgson, R.D. Organic Chemistry of Explosives, John Wiley & Sons, England, 2007
- Mulliken, R.S.J. Chem. Soc. 1950, 72, 4493
CrossRef - Parini, V.P. Chem. Rev. 1962, 31, 408
CrossRef - Buncel, E., Norris, A.R., Russell, K.E.Q. Chem. Soc. 1968, 22, 12
CrossRef - Foster, R. Organic Charge transfer complexes, Academic Press, London 1969
- Mulliken, R.S. Am. Chem. Soc. 1952, 74, 811
CrossRef - Russell, G.A., Janzen, E.G. Am.Chem.Soc. 1900, 84, 453
- Jackson, C.J., Gazzalo, F.H. Am.Chem.Soc. 1902., 23, 376
CrossRef - Akhavan, J. ” The Chemistry of Explosives” Royal society of chemistry, Great Britain, 2nd ed., 2004
- Tripathi, K.D. “ Essentials of Medicinal Pharmacology”, 6 th ed., Japyee Brothers Medicals publishers, Chennai, 2009
- Molisz, J., Kaeser, A., Kaufer, R., Lautenbatch, H., polley, E., Hoffmann, M. Patent number WO2001032786, 2001
- Kaler, G.,k Threfall, C.J., Basava, C., Okum, I. Patent number US 2001-315276P, 2001
- Gregory, P.F., Clive, E., O’ Shaughnessy, H.A. Patent number WO2002072715, 2002
- Rezende, M.C., Flores, P., Guerrero, J., Villarroel, L. Acta. 2004, 60A, 1637
CrossRef - Jackson, C.J., Gazzalo, F.H. Am.Chem.Soc. 1902, 23,376
CrossRef - Mulliken, R.S. Am. Chem. Soc. 1952, 74, 811
CrossRef - Russell, G.A., Janzen, E.G. Am.Chem.Soc. 1900, 84, 453
CrossRef - Black .J.A., Evans, M.J.B., Russel, K.E. J.Chem.. 1996, 44,119
CrossRef - Meisenheimer, J. Justus liebigs Ann. Chem . 1902, 323,205
CrossRef - Bunnett, J.F., Zahler, R.E. Rev. 1951, 49,273
CrossRef - Bunnett, J.F. Rev. 1958, 12, 1
CrossRef - Sauer, J., Huisgen, R. Chem. 1960, 72, 294
CrossRef - Ross, S.D. progress Phys. Org. Chem. 1963, 1, 31
CrossRef - Buncel, E., Dust, J.M., Terrier, F. Rev. 1995, 95, 2261
CrossRef - Makosza, M., Wojciechowski, K. Rev. 2004, 104, 2631
CrossRef - Iluminada, G., Gonzalo, G., Jordi, M. Chem. Common. 2002, 22, 2638
- Makosza, M., Lemek, T., Kwast, A., Terrier, F. J. Org. Chem. 2002, 67, 394
CrossRef - Isanbar, C., Emokpae, T.A., Crampton, M.R., Chem. Soc. Perkin Trans 2, 2002
- Crampton, M.R. Phy. Org. Chem. 1969, 7 ,211
CrossRef - Strauss, M. Chem. Rev. 1970, 70,667
CrossRef - Terrier, F. Rev.1982, 82, 77
CrossRef - Silverstein, R.M., Webster, F.X. Spectrometric identification of organic compounds, Chem. Educ. 1962, 39, 11, 546 John Wiley and Sons, New York,2004, P.130
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









