Synthesis and the Study of Bioefficacy of Schiff Base Ligand Decorated by Pyrazolone Moiety
Anuradha Rajasekaran1*, Selvan Abraham2, Sasikala Mohanasundaram3 and Pirakathiswaran Gurusamy1
1Research and Development Centre, Bharathiar University, Coimbatore, India -641046.
2Department of Chemistry, The American college, Madurai, India – 625002.
3Department of Chemistry, Dr. M.G.R Educational and Research Institute, Chennai, India – 600095.
Corresponding Author E-mail: ranuradha18@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/360205
Article Received on : 13 Feb 2020
Article Accepted on : 20 Apr 2020
Article Published : 30 Apr 2020
The study reports synthesize of novel Schiff base ligand decorated with pyrazolone moiety using 4-Aminoantipyrine phosphate and 5-Bromosalicylaldehyde. Structural characterization of the synthesized pyrazolone moiety was carried out using 1H-NMR, Mass Spectroscopy, elemental analysis with EDAX, UV-Vis spectroscopy, Fourier Transform Infrared Spectroscopy (FTIR) and single crystal X-Ray diffraction. Novel Schiff base ligand was tested for its bioefficacy, which showed having the properties like antimicrobial against Escherichia coli (MTCC433), Staphylococcus aureus (MTCC3160), antifungal against Aspergillus niger (MTCC2208) Candida albicans (MTCC3160), antioxidant and anti-inflammatory activities. Cytotoxicity study was carried out on Hep G2 cell lines and Vero cell lines, which revealed the anticancer activity of the Schiff base ligand. Molecular docking interaction of ligand towards enzymes like DNA gyrase of E.coli, S. aureus showed good binding energy. The optimum synthesizing temperature of Schiff base ligand was found to be at 30°C, and the synthesised ligand was found to be biologically effective.
KEYWORDS:Anti-Cancerous Activity; Bioefficacy; Docking Studies; Hep G2 Cell Line Studies; Schiff base Ligand
Download this article as:| Copy the following to cite this article: Rajasekaran A, Abraham S, Mohanasundaram S, Gurusamy P. Synthesis and the Study of Bioefficacy of Schiff Base Ligand Decorated by Pyrazolone Moiety. Orient J Chem 2020;36(2). |
| Copy the following to cite this URL: Rajasekaran A, Abraham S, Mohanasundaram S, Gurusamy P. Synthesis and the Study of Bioefficacy of Schiff Base Ligand Decorated by Pyrazolone Moiety. Orient J Chem 2020;36(2). Available from: https://bit.ly/35qoR7i |
Introduction
Schiff’s bases are one of the most novel non-transitional metal complexes owing to its own appropriate properties, application, and employment in each theoretical and sensible aspect. This in-depth application of Schiff base advances involving element and chemical element Donor ligand attracts the chemist to synthesis and characterize the new pyrine based mostly on Schiff bases and its auriferous complexes.1 Coordination polymers, typically observed as Metal Organic Frameworks (MOFs), are presently attracting a high level of contemplation in chemical science. This MOF design has witnessed a vast development as a result of their intriguing structures and potential properties.2 Such MOFs are often arrived by the condensation reaction of carbonyl compounds and amines, which leads to the formation of Schiff bases, namely imine compounds.3 The developed Schiff base compound has received an awesome attention, and additionally, these Schiff base macrocyclic complexes remarkably play the role of causation substrate chirality, enhancing solubility, catalyst, etc.4
There are widespread applications of the Schiff base
ligands in the field of biological, analytical, clinical and
industrial aspects. Among these, the heterocyclic Schiff base ligands advanced do have vital interest as a result of pharmacological properties
and has evolved in production of therapeutic agents. The antibacterial and antifungal
studies were allotted by
in vitro methodology against Escherichia coli, Staphylococcus aureus, and Aspergillus niger, Candida
albicans so as to access
their potential.5
Cancer seems to be a
serious reason for morbidity
and mortality and runs within the prime three causes of death worldwide, particularly within
the developed countries. Therapy is one of all the potential treatments for
prolonging the patient’s life; however, as a
result of these, medicines travel
throughout the body; they will have an effect on normal cells
also. So as to
develop associate degree alternate
system, recent research workers focus
on the malignant tumor studies. This malignant tumor study involves learning of several biological assays. It
needs the mensuration of living and/or proliferate learning class cells. Toxicity assays are widely employed in in-vitro pharmacological medicine studies.
Among the various cytotoxicity assays available,
this study reported MTT assay method for evaluation of anti-cancerous and
toxicity activity of the Schiff based ligand6. Human hepatoblastoma
derived Hep G2, a prominent equivalent of hepatotoxicity studies proved to be
effective screening and monitoring method to study toxicity and metabolism
activity of the pyrazolone derivatives.7
The antioxidant has been incontestable that free radicals will harm proteins, lipids and deoxyribonucleic acid of bio-tissues, resulting in an inflated rate of cancer.8 Luckily, antioxidants will forestall this harm, thanks to their radical scavenging activity.9 The insight of this focuses on the bioefficacy of the synthesized matter.10 In this context, our interest is targeted for a substantial preparation and characterization of latest, stable, inert and non-toxic Schiff base matter from 4-Aminoantipyrine phosphate and 5-Bromosalicylaldehyde and also the bioefficacy studies to evaluate the specified biological activity.
Materials and Methods
Reagents and chemicals used in the study were of analytical grade. 4-amino-1,5-dimethyl-2-phenyl- 1,2-dihydro-3H-pyrazol-3-one and 5-Bromo-2-hydroxybenzaldehyde were procured from Merck. Ligand was prepared using the commercial solvents which was distilled before its use. Gentamycin, Acridine orange and amphotericin B were purchased from Sigma and MTT (Thiazolyl blue tetrazolium bromide) was procured from Invitrogen.
Synthesis of Ligand
The Schiff base ligand was prepared as follows. An ethanolic solution of 4-amino-1,5-dimethyl-2-phenyl-1,2-dihydro-3H-pyrazol-3-one (0.500 g, 0.002 mol) and ethanolic solution of 5-Bromo-2-hydroxybenzaldehyde (0.494 g, 0.002 mol) was refluxed for 6 h. The reaction mixture was refluxed for three different temperatures (25°C, 30°C and 35°C), and the optimum temperature for the pyrazolone based Schiff base was studied.11, 12
Spectrum Analysis
The composition percentage of the elements of the ligand was determined using elemental analyzer Thermo Finnigan FLASH 1112 CHNS analyzer. FTIR analysis of the ligand was done with Perkin Elmer Spectrophotometer (Spectrum GX model). 1H NMR spectra was determined with a Brucker WM (400 MDCXHz) and Deuterated Dimethyl sulfoxide as standard reference. GC mate benchtop double focusing Mass Spectrometer was used to recorder the Mass spectrum of the compound. Systronics 2201 double beam spectrophotometer depicted the Electronic absorption spectra of the ligand. Single crystal X-ray diffraction analysis and Melting point of Schiff base ligand was performed on a Brucker Apex II CCD diffractometer Veego DS model apparatus respectively. Spectral data were correlated using APEX II software with SADABS extension of APEX for correction of spectrum.
Antimicrobial Studies
The in vitro antibacterial activity of the compound against gram-negative E. coli (MTCC433), gram-positive S. aureus (MTCC3160) was carried out using Muller Hinton agar media. Agar Disc diffusion method was carried out to analyze the antibacterial activity, with streptomycin (30 µg/mL) as the standard. The uniform size wells (10 mm diameter) were made in the agar plates and was loaded with 15, 20, and 25 µl of the test samples and incubated for 24 hrs at 27°C. Inhibition zone of the organism was evaluated by measuring the clear zone of inhibition formed around each well.13
The newly synthesized ligand was also screened for its antifungal property against A. niger (MTCC2208) and C.albicans (MTCC3160) on Sabouraud agar Medium. After swabbing with the test organism, the agar plates were bored using sterile cork to well uniform pores of 10 mm diameter. Test compound was added with varying volume of 15, 20, and 25 µl. Finally the plates were kept at 37°C for 3 days for incubation and zone of inhibition were measured for determining the antifungal activity.14
In Vitro Assay for Anticancerous Activity
Cell Line and Culture
Vero cell line was obtained through Pune, NCCS – National Center for Cell Science Centre. The cells were maintained at a humidified atmosphere of 50 μg / mL CO2 at 37 ° C, in DMEM supplemented with 10 % FBS and antibiotics such as penicillin and streptomycin of concentration 100 μg/mL.
Vero Cell Line MTT Assay
Vero cell line of 1 × 105 /well of cell density were seeded in a 24 well plate and incubated in 37°C with 5% CO2. After the cell reached its confluence point, samples were added at different concentration and the plates were held for 24 hours of incubation. After incubation period, the sample was drained and washed with DMEM (Dulbecco’s Modified Eagle Medium) without serum (pH 7.4). 100 microliters of MTT (Thiazolyl blue tetrazolium bromide) of stock concentration 5 mg/mL was added to each well and incubated for 4 hours. Wells inoculated with samples were rinsed with 1 mL of DMSO and transferred to UV-spectrophotometer for measuring absorbance at 570 nm. Pure DMSO was kept as blank and IC50 value of the test sample was graphically determined. (Concentration of sample for 50% inhibition) 15
Cytotoxicity Assay on Hep G2 cell lines:
Hep G2 cells grown in RPMI media added 10% fetal bovine serum along with antibiotics like penicillin were procured from NCCS. (National Centre for Cell Science, Pune). Cells were maintained in 5% CO2 at temperature of 37°C until the required confluence is developed. Cells of density 5 × 103 cells/well were seeded in 96 well plates containing 200 µl of RPMI with 10% FBS and incubated for 24 hours. After the incubation period, supernatant was drained and the cells were inoculated with test compound of varying concentration and were allowed to react for 48 hours. Then the test compound were drained out and 100 µl of DMSO containing MTT (10 µl, 5 mg/mL) were added and incubated at 37°C for 4 hours. Finally the culture media with MTT in DMSO were taken out from the well plate and absorbance were measured at 595 nm.
The cell viability was calculated using the following formula:
Cell viability (%) = (Average test OD/Control OD) × 100
Antioxidant Activity – DPPH Assay
DPPH is one of the most stable free radical which will not allow any dimer formation of molecule owing to its affinity towards delocalization of free electrons over the free ionic state of the test compound. The shift in electronic state causes the formation of deep violet color which can be measured has an alteration in absorbance at 517 nm. Initially 0.1 mM stock solution of DPPH was prepared in methanol. Then 800μl of DPPH solution was added to 200μl of test compound in varying concentration ranging from 10 to 1000μg/mL. The reaction mixture were shaken vigorously and were incubated at 37°C in a dark room for 30 min. Finally formation of violet color was measured at 517 nm by using spectrophotometer and change in absorbance between blank (without test compound) and test compound of different concentrations were plotted as graph to estimate the free radical scavenging activity of the test compound.16, 17
Inhibition of Inhibition (%) = (ABlank − Asample/ABlank) × 100
Where: ABlank = absorbance of the reaction mixture without test compound; Asample = absorbance of the tested compounds
In Vitro Anti-Snflammatory Studies
Stock solution of the test compound with the concentration of 10 mg/mL was prepared using methanol. Test compound was dissolved in methanol in dilutions of 100, 200, 300, 400, and 500 µg/mL. 0.2% stock solution of bovine serum albumin (BSA) was prepared with Tris buffer. To 50 µl of the each dilution of the test compound, 5 mL of BSA was added and the samples were incubated at 37°C for 20 min and incubated again at 72°C for 5 min. Finally the samples were allowed to cool for 5 min at room tenpertaure and the absorbance was measured at 660 nm. With the obtained OD values the anti-inflammatory reaction was calculated by the percentage of inhibition of protein denaturation by given formula:
Inhibition of denaturation (%) = (Acontrol − Asample/Acontrol) × 100
where: Acontrol = absorbance of the reaction mixture without test compound; Asample = absorbance of the tested compounds.
In-Silico Analysis using Molecular Docking
Target Protein and Ligand Preparation
The crystal structure of target proteins such as DNA gyrase from E.coli (1AB4) and dehydrosqualene synthase from S. aureus (3ACX) were retrieved from the database of Protein Data Bank (RCSB). The Chem 3D ultra 11.0 software was used to construct the three-dimensional structures of the current syntheszied Schiff based ligand. Kollmann charges, polar hydrogen bonds were added to the protein, and simultaneously all bound water and ligands were removed.
Protocol of Docking Studies
Auto dock version 4.0 software was used for the automated docking study of the synthesized Schiff based ligand. The auto grid, a component of the auto dock, was used to compute the grid maps with the interaction energies depending upon the macromolecule target of the docking study. The grid center was placed on the active target site region of the enzyme. Then binding free energy of the inhibitors was evaluated using automated docking studies. The best conformations search was done by adopting genetic algorithm with local search (GA-LS), method. 50 independent runs of docking were carried out with Auto-Dock Tool. Root mean square (RMS) tolerance of 2.0 Å was performed using structures generated after completion of docking via cluster analysis. Molecular graphics and visualization were performed with the UCSF Chimera package.18, 19
Results and Discussion
The synthesized Schiff base ligand appeared as a yellow crystal, which was dissolved in ethanol to form yellow precipitate and again recrystallized to maintain the purity of the sample. Condensation reaction between 4-amino-1,5-dimethyl-2-phenyl-1,2-dihydro-3H-pyrazole-3-one and 5-Bromo-2-hydroxybenzaldehyde were carried out at different temperature (25°C, 30°C and 35°C). The yield was observed as 0.60, 0.75 and 0.65 g respectively. The results depicted that the synthesized ligand appeared to be stable at 30°C, which was taken as the optimum temperature for the synthesis of ligand. The melting point of the ligand is measured as 189–194°C. Elemental analyses of ligand (Table 1) and other spectral studies were used to predict the structure of the synthesized ligand (Figure 1).
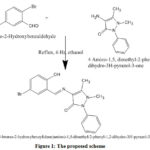 |
Figure 1: The proposed scheme |
Table 1: Elemental analysis and physical properties of ligand – Theoretical (% Calculated)
|
Compound |
Empirical Formula |
Color |
Yield (%) |
C |
H |
N |
|
Ligand |
C18H16BrN3O2 |
Yellow |
85 |
55.97 (55.94) |
4.18 (4.14) |
10.88 (10.82) |
FT- Infrared Spectrometry Analysis
The spectrum analysis of the Schiff base ligand which was carried out in the range of 4000–400 cm−1, helped in elucidating the structure analysis by revealing the functional groups present in the ligand (Figure 2). The absence of NH2 stretching vibrations was confirmed by no absorption peaks at the range of 3400–3300 cm−1. In general the peak at 1631.00 cm−1 which corresponds to the presence of C=O (an aromatic ring) and C=N bonds. The absence of NH2 group and peak formation of C=N group corresponds to the effective condensation of the reactants, by removal carbonyl oxygen of 5-Bromo-2-hydroxybenzaldehyde and the amino group of 4-amino-1,5-dimethyl-2-phenyl- 1,2-dihydro-3H-pyrazole-3-one, leading to imine group C=N. Other peaks at 1477.27 cm−1 correspond to the C=C of the aromatic ring. CH group presence was confirmed by the in-plane bending at 1120.76 cm−1. N-CH3 wagging was due to peak presence at 698.78 cm−1.20 3.2 1H-NMR
 |
Figure 2: FTIR spectra of the ligand |
The signal at 9.61 ppm corresponds to the CH=N proton and absence of proton signal for free NH2 groups at 2.99 ppm provides the evidence supporting the condensation and formation of Schiff base ligand. The multiplets signal between 6.86 ppm and 7.62 ppm formed due to the presence of the phenyl proton. The spectral display (Figure 3) at 3.334–3.328 ppm corresponds to the three hydrogen atoms in C-CH3 group 21. 1H-NMR spectral data of 4-((5-bromo-2-hydroxybenzylidene)amino)-1,5-dimethyl1-2-phenyl-1,2-dihydro-3H-pyrazol-3-one: δ 2.31 (3H, s), 3.64 (3H, s), 6.81 (1H, dd, J = 8.4, 0.4 Hz), 7.17 (1H, dd, J = 8.4, 1.6 Hz), 7.34 (1H, tt, J = 7.6, 1.3 Hz), 7.43 (2H, dddd, J = 8.2, 1.5, 1.3, 0.5 Hz), 7.60 (2H, dddd, J = 8.2, 7.6, 1.5, 0.5 Hz), 7.75 (1H, dd, J = 1.6, 0.4 Hz), 8.54 (1H, s).
 |
Figure 3: 1H NMR of the ligand |
Spectral Analysis – Molecular Mass
The molecular ion peak observed at 398 m/z obtained from ESI spectrum analysis (Figure 4) of Schiff base ligand, correlates to the molecular weight 386.25 of the monoclinic ligand. The empirical formula of Schiff base ligand C18H16BrN3O2 was suggested using data from analytic and elemental analysis. Other prominent peaks in the mass spectrum corresponds to the sub-structural moieties of the Schiff base ligand. Molecular ion peak observed at 189.16 m/z corresponds to parent compound 4-amino-1,5-dimethyl-2-phenyl-1,2-dihydro-3H-pyrazole-3-one. Similarly ionic peaks at 122.10 m/z and 202.03 m/z correlates to the sub-structural moieties Diazane, Methyl phenyl and acetyl phenyl pyrazolidine respectively.
 |
Figure 4: Mass spectra of the ligand |
Electronic spectra and single crystal XRD studies
The spectral studies were analyzed by dissolving the test compound in Deuterated DMSO. The absorption band at the 321, 341, and 426 nm were endorsed to the π → π*, n → π* shifts of the Schiff based ligand.22 Bands absorption in the range of 235–426 nm are mostly detected due to π → π*, n → π* electronic shifts of C=N, a benzene ring or it can be attributed to the involvement of either metal to the ligand, ligand–metal electron transfer originating due to π interactions. The electronic spectra of the Schiff based ligand revealed a band at 477 nm, assigned to 2B1g→2A1g transitions supports the characteristics of square planar geometry around the metal ion.22 Further supported by magnetic susceptibility study, magnetic moment value was found to be 1.83/1.9 BM. Structural analysis of the ligand using Single crystal XRD is a precise way to confirm structure; the raw diffraction pattern file rendered from XRD analysis was used to solve the structure of Schiff base ligand. Initially three dimensional coordinates (hkl) were generated from the diffractogram (Figure 5a). Then, using the miller indices (hkl) the three dimensional structure of Schiff base ligand was plotted. Hence, single crystal XRD investigations confirmed the predicted structure ((E)-4-((5-bromo-2-hydroxybenzylidene)amino)-1,5-dimethyl1-2-phenyl-1,2-dihydro-3H-pyrazol-3-one) (Figure 5).
 |
Figure 5a: XRD pattern of the ligand (solved) |
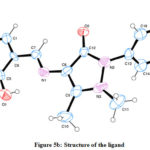 |
Figure 5b: Structure of the ligand |
Antimicrobial Activity of Schiff Based Ligand
Evaluation of antibacterial and antifungal potential of the Schiff based ligand was carried out using well diffusion method. Antibacterial activity of the test compound was screened against S. aureus (gram-positive bacteria) and E.coli (gram-negative bacteria) with streptomycin as standard drug. Similarly for antifungal activity, test compound was screened against A. niger, and C.albicans with ketoconazole as the standard drug. The biological activity was studied for 24 hrs (bacterial plates) and 3 days (fungal plates) by measuring the zone of inhibition formed. (ZOI).23, 24
The results depicted in Table 2 shows that the ligand has shown distinct antibacterial activity against both S. aureus (gram-positive) and E.coli (gram-negative bacteria). The ZOI observed at a different concentration showed highest zone of 22 mm (Figure 6b) at 50 µg/mL which was greater to the zone of inhibition measured for the standard at 24 h study against E. coli. Against S. aureus, ligand showed a maximum zone of inhibition of about 27 and 16 mm (Figure 6a) at 50 and 25 µg/mL, respectively. The ligand had better activity than the standard drug streptomycin at 30 µg/mL, which had a zone of inhibition of 17 mm (Figure 6c) and 14 mm (Figure 6d) against Staphylococcus aureus and Escherichia coli respectively.25
 |
Figure 6: Antibacterial activity of the ligand |
Table 2: Antibacterial activity of ligand
|
Compound |
Zone of inhibition (mm) |
|||
|
Streptomycin |
Ligand |
|||
|
Concentration |
30 µg/mL |
10 µg/mL |
25 µg/mL |
50 µg/mL |
|
Staphylococcus aureus |
14 |
13 |
16 |
27 |
|
Escherichia coli |
17 |
12 |
14 |
22 |
Antifungal activity is given in Table 3 against A. niger, C. albicans. The Schiff base ligand had moderate activity against the A. niger, having Zone of 25 mm (Figure 7a) at 50 µg/mL concentration and Zone of 18 mm (Figure 7b) was depicted for ligand of concentration 50 µg/mL against C. albicans. On comparing the Zone of 30 mm and 36 mm by standard antifungal drug Ketoconazole at 30 µg/mL, it was clear that Schiff base ligand has potent antifungal nature. Hence, it was evident that Schiff base ligand possesses good antibacterial and antifungal activity against the tested organisms.26, 27
 |
Figure 7: Antifungal activity of the ligand |
Table 3: Antifungal activity of ligand
|
Compound |
Zone of inhibition (mm) |
|||
|
Ketoconazole |
Ligand |
|||
|
Concentration |
30 µg/mL |
10 µg/mL |
25 µg/mL |
50 µg/mL |
|
Aspergillus niger |
30 |
11 |
14 |
25 |
|
C.albicans |
36 |
13 |
16 |
18 |
Antioxidant Activity
DPPH free radical scavenging assay is one of the most extensively used methodology for the purpose of evaluation of antioxidant acivity of any organic or inoganic test compound. The mechanism is that when the sample with antioxidant property reacts with DPPH radical, it causes a decrease in absorbance of DPPH due to the sample’s scavenging activity by hydrogen donation, which can be visually noticed by color from purple to yellow (antioxidant property). The Schiff based ligand showed the scavenging activity of about 90.45% at 1 mg/mL concentration (Table 4).
Table 4: The Antioxidant activity of ligand using DPPH assay method
|
S. No. |
Sample |
Concentration (µg/mL) |
OD (nm) (blank OD = 0.733) |
DPPH activity (%) |
|
1 |
Ligand |
1000 |
0.070 |
90.45 |
Cytotoxicity Assay on Hep G2 Cell Lines
The study was focused to determine the ability of the ligand to be a potent anticancer agent. After evaluation of the antioxidant property of the Schiff based ligand, it prompted us to investigate the in-vitro anticancerous activity of the ligand against Hep G2 (Figure 8) (hepatocarcinoma) and Vero cell lines (Figures 9) using MTT assay.
 |
Figure 8: Cytotoxicity study of the ligand against Hep G2 Click here to View Figure |
Table 5: Anti-cancerous activity of the ligand against Hep G2 cell lines
|
Compounds |
Sample |
Positive control |
|||||
|
1 ng |
10 ng |
100 ng |
1 µg |
10 µg |
100 µg |
10 ng |
|
|
OD – 1st Trial (nm) |
0.567 |
0.517 |
0.424 |
0.307 |
0.237 |
0.206 |
0.432 |
|
OD – 2nd Trial (nm) |
0.572 |
0.509 |
0.411 |
0.311 |
0.245 |
0.199 |
0.419 |
|
Avg (nm) |
0.5695 |
0.513 |
0.4175 |
0.309 |
0.241 |
0.2025 |
0.4255 |
|
1-t/c |
0.21826 |
0.29581 |
0.42691 |
0.57584 |
0.66918 |
0.72203 |
0.41592 |
|
%Inhibition of cancer cells |
21.8257 |
29.5813 |
42.6905 |
57.5841 |
66.9183 |
72.2032 |
41.5923 |
Vero cell lines were used as a control cell line to measure the safety of the Schiff based ligand. The Tables 5 and 6 give the information that the ligand had anti-proliferative activity against Hep G2 cell line and Vero cell line respectively 28, 29. Both figure 8 and figure 9 gives the clear illustration regarding the proliferation of that particular cell line under the influence of synthesized Schiff base ligand at various concentrations.
 |
Figure 9: Cytotoxicity effect of Sample on Vero cell line |
Table 6: MTT assay (Cytotoxicity) of Schiff base ligand against Vero cell line
|
S. No. |
Concentration (µg/mL) |
Absorbance |
Cell viability (%) |
|
1 |
1000 |
0.417 |
49.46 |
|
2 |
500 |
0.466 |
55.27 |
|
3 |
250 |
0.528 |
62.63 |
|
4 |
125 |
0.585 |
69.39 |
|
5 |
62.5 |
0.648 |
76.86 |
|
6 |
31.2 |
0.704 |
83.51 |
|
7 |
15.6 |
0.767 |
90.98 |
|
8 |
7.8 |
0.822 |
97.50 |
|
9 |
Cell control |
0.843 |
100 |
In vitro Anti-inflammation Activity of Schiff Ligand
In vitro anti-denaturation of protein study results depicted in Table 7, it can be observed that Schiff based ligand possesses anti-inflammatory activity and it increases as the concentration of the ligand increases, similarly IC50 value was found to be at 195.62 µg/mL. A potent anti-inflammatory agent should have more than 20% inhibition of protein denaturation,30 which leads to the observation that Schiff based ligand possessed good anti-inflammatory activity even at 100 µg/mL.
Table 7: Anti-inflammatory activity of ligand
|
S. No. |
Sample Concentration (µg/mL) |
% Inhibition |
|
1 |
100 |
35.94 |
|
2 |
200 |
55.47 |
|
3 |
300 |
61.72 |
|
4 |
400 |
71.88 |
|
5 |
500 |
90.63 |
|
6 |
IC50 |
195.62 |
Auto-Docking of the Schiff Based Ligand
The docking results of the schiff based ligand are summarized in Tables 8 and 9. The binding energy (Kcal/mol) and inhibition constant (Ki) and hydrogen bonds formed were used to evaluate binding affinity of the inhibitors.
Table 8: Auto docking of ligand into DNA gyrase from E.coli (1AB4)
|
Compound Name |
Binding Energy (kcal/mol) |
Ki |
Hydrogen bond interaction |
|
|
Hydrogen Donor |
Hydrogen Acceptor |
|||
|
Ligand |
−2.65 |
11.36 mM |
1ab4: LYS 42 |
Ligand |
Table 9: Auto docking of ligand into Staphylococcus aureus (3ACX)
|
Compound Name |
Binding Energy (kcal/mol) |
Ki |
Hydrogen bond interaction |
|
|
Hydrogen Donor |
Hydrogen Acceptor |
|||
|
Ligand |
−4.59 |
.42987 mM |
3acx: ASN 168 3acx: TYR 41 |
Ligand |
From the binding model, the schiff based ligand (Figure 10) was most potent towards the active catalytic site binding of the Staphylococcus aureus, via two hydrogen bonds formation between the compound of interest and amino acid residues ASN 168 (2.713 Å), TYR 41 (2.853 Å). Similarly, the same compound showed a less binding affinity towards DNA gyrase of E.coli(Figure 11) forming one hydrogen bond between the compound and amino acid LYS 41 (2.997 Å).
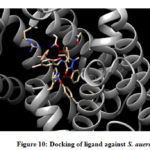 |
Figure 10: Docking of ligand against S. auerus |
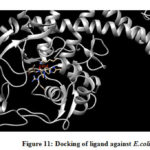 |
Figure 11: Docking of ligand against E.coli |
Conclusion
Finally, we report a synthesis of a novel compound (E)-4-((5-bromo-2-hydroxybenzylidene)amino)-1,5-dimethyl1-2-phenyl-1,2-dihydro-3H-pyrazol-3-one) , which had the optimum synthesis at the temperature of 30°C. The structure of the ligand was proposed on the basis of elemental analyses, FTIR, UV-Visible, 1HNMR, Mass spectra. The bio-efficacy studies performed for the synthesized structures clearly indicate that ligand had good antibacterial properties, which was also supported by autodock studies of the ligand. Schiff based ligand in the biological screening against the cancer cell Hep G2 cell line showed as potent anti-tumor with less cytotoxic when screened against Vero cell lines. The anti-oxidant and anti-inflammatory activity were also found to be effective, proving the fact that our synthesized Schiff base are biologically active and highly potential pyrazolone moiety and holds a promising future as an excellent pharmacophore for drug development purposes. Future studies will be carried to study in detail about the bio efficacy of the materials as a potent drug for Cancer and other comorbid conditions.
Acknowledgements
One of the authors, Anuradha is thankful to the Management, Principal and Head of the Department of Chemistry, Vel Tech High Tech Dr. Rangarajan Dr. Sakunthala Engineering College, Chennai for providing necessary research facilities. They are grateful to SAIF, IITM for recording NMR and Mass spectra. They also extend their gratitude towards Mahatma Gandhi University, Kottayam for the X-Ray crystallographic facility.
Supplementary Information (SI)
Crystallographic data for the structural analyses of ligand is available in the supplementary information. Supplementary Information is available at www.ias.ac.in/chemsci.
References
-
- Chellaian, J .D.; Jijo, J. Acta Part A. 2014, 118, 624-631.
- Nayan, H. B.; Pratik, K. T.; Pankaj, M. S.; Vrajlal, K. G.; Viresh, H. S. Saudi Chem. Soc. 2015, 21(5), 517-527.
- Muhammad, A.; Itrat, A.; Nighat, A.; Rashad, M.; Ajaz, H.; Tanveer, H. B.; Muhammed, T. H.; Haq, N.; Muhammed, K. Saudi Chem. Soc. 2015, 19(3), 322-326.
- Mohammad, S. A.; Jung-Hyun, C.; Dong-Ung, L. Med. Chem. 2012, 20(13), 4103-4108.
- Wojciech, S.; Bohdan, K.; Anna, S. C.; Beata, K.; Eugeniusz, G.; Dorota, Z.; Anna, W.; Tadeusz, O. Acta Part A. 2013, 109, 47-54.
- Dehn, F.; White, C.M.; Conners, D. E.; Shipkey, G.; Cumbo, T. A. Biol. Anim. 2004, 40(5-6), 172-182.
- Yoshitomi, S.; Ikemoto, K.; Takahashi, J.; Miki, H.; Namba, M.; Asahi, S. Toxicol In Vitro. 15, 2001, 15(3), 245-256.
- ; Junod, A. F. Eur. J. Cancer. 1996, 32(1), 30-38.
- Yu, Chi.; Taizoon, C.; Hans, H. C. C.; U-Ser, J.; Tsang-Lang, L.; Long, Y. C. Fullerene Nanotechnol. 2000, 165-183.
- Basaar, O.; Fatema, S.; Mohsin, M.; Farooqui, M. J. Pharm. Biol. Chem. Sci. 2017, 8(3), 1857-1866.
- Syed, A. M.;Nighat, F. Pharmacognosy Magazine. 2015, 11(Suppl 1), S123.
- Raman, N.; Sobha, S.; Selvaganapathy, M.; Mahalakshmi, R. Acta Part A Mol. Biomol. Spec. 2012, 96, 698-708.
- Anant, P.; Bibhesh, K. S.; Narendar,; Devjani, A. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2010, 76(3-4), 356-362.
- El-Sonbati, A. Z.; Diab, M. A.; El-Bindary, A. A.; Abou-Dobara, M. I.; Seyam, H. A. Acta Part A Mol. Biomol. Spectros. 2013, 104, 213-221.
- Mosmann, T. Immunol. Method. 1983, 65(1-2), 55-63.
- Molyneux, P. J. Sci. Technol. 2004, 26(2), 211-219.
- Kedare, S. B.; Singh, R.P. J. Food Sci. Technol. 2011, 48(4), 412-422.
- Saveg,; Shrish, K. P.; Vinay, K. S.; Yugal, G.; Ajay, K.; Sukh, M. S.; PLOS ONE. 2017, 12(5).
- Abdel, M.; Adam, A. Acta Part A Mol. Biomol. Spectrosc. 2013, 104, 1-13.
- [20] Raman, N.; Sobha, S.; Selvaganapathy, M.; Mahalakshmi, R. Acta Part A Mol. Biomol. Spectrosc. 2012, 96, 698-708.
- Amit, K. S.; Sulekha, C. Acta Part A Mol. Biomol. Spectrosc. 2011, 81(1), 424-430.
- Raman, N.; Sobha, S. Acta Part A Mol. Biomol. Spectrosc. 2012, 85(1), 223-234.
- Anitha, C.; Sheela, C. D.; Tharmaraj, P.; Sumathi, S.; Acta Part A Mol. Biomol. Spectrosc. 2012, 96, 493-500.
- Yogesh, S. P.; Khyati, D. P.; Hasmukh, S. P. Saudi Chem. Soc.2012, 20, S300-S305.
- Tudor, R.; Elena, P.; Catalin, M.; Rodica, G.; Nicolae, S.; Aurelian, G. 2011, 30(1), 154-162.
- Joseph, J.; Nagashri, K.; Ayisha, B. R. G. Saudi Chem. Soc.2013, 17(3), 285-294.
- Samir, A.; Hanan, F. E.H.; Dahshan, A. Mol. Struct. 2010, 983(1-3), 32-38.
- Fugu, M. B.; Ndhahi, N. P.; Paul, B. B.; Mustapha, A. N. Chem. Pharm. Res. 2013, 5(4), 22-28.
- Renau, T. E.; Kennedy, C.; Ptak, R. G.; Breitenbach, J. M.; Drach, J. C.; Townsend, L. B. Med. Chem. 1996, 3470-3476.
- DRĂGAN, M.; STAN, C. D.; Iacob, A.; PROFIRE, L. 2016, 64(5), 717-721.

This work is licensed under a Creative Commons Attribution 4.0 International License.









