Triterpenoids from the Bark of Aglaia Glabrata and Their In Vitro Effects on P-388 Murine Leukemia Cells
Desi Harneti1, Asep Supriadin2, Rani Maharani1,3, Nurlelasari1, Tri Mayanti1, Ace Tatang Hidayat1,3, Risyandi Anwar4, Unang Supratman*1,3 , Khalijah Awang5 and Yoshihito Shiono6
, Khalijah Awang5 and Yoshihito Shiono6
1Department of Chemistry, Faculty of Mathematics and Natural Sciences, Universitas Padjadjaran, Jatinangor 45363, Sumedang, Indonesia.
2Department of Chemistry, Faculty of Science and Technology, Sunan Gunung Djati Islamic University, Bandung 45262, Indonesia.
3Central Laboratory, Universitas Padjadjaran, Jatinangor 45363, Sumedang, Indonesia.
4Department of Pediatric Dentistry, Faculty of Dentistry, Muhammadiyah University, Semarang 50273, Central Java, Indonesia.
5Department of Chemistry, Faculty of Science, University of Malaya, Kuala Lumpur 59100, Malaysia.
6Department of Food, Life, and Environmental Science, Faculty of Agriculture,Yamagata University, Tsuruoka, Yamagata 997-8555, Japan.
Corresponding Author E-mail: unang.supratman@unpad.ac.id
DOI : http://dx.doi.org/10.13005/ojc/350114
Article Received on : 25-10-2018
Article Accepted on : 03-01-2019
Article Published : 31 Jan 2019
Four dammarane-type triterpenoids, dammardienon (1), aglaiabbreviatin E (2), dammar-20,25-dien-3b,24-diol (3) and dammar-24-en-3b,20-diol (4) were isolated from methanolic extract of the bark of Aglaia glabrata. The structures of all triterpenoids were elucidated by 1D-, 2D-NMR, and comparison with previously reported data. All triterpenoids were applied into in vitro bioassay against P-388 murine leukemia cell. Dammar-24-en-3b,20-diol (4) has cytotoxic activity with IC50 value of 9.45 mM towards P-388 murine leukemia cell lines.
KEYWORDS:Aglaia Glabrata; Dammarane-Type Triterpenoid; Meliaceae; P-388 Murine Leukemia Cell
Download this article as:| Copy the following to cite this article: Harneti D, Supriadin A, Maharani R, Nurlelasari N, Mayanti T, Hidayat A. T, Anwar R, Supratman U, Awang K, Shiono Y. Triterpenoids from the Bark of Aglaia Glabrata and Their In Vitro Effects on P-388 Murine Leukemia Cells. Orient J Chem 2019;35(1). |
| Copy the following to cite this URL: Harneti D, Supriadin A, Maharani R, Nurlelasari N, Mayanti T, Hidayat A. T, Anwar R, Supratman U, Awang K, Shiono Y. Triterpenoids from the Bark of Aglaia Glabrata and Their In Vitro Effects on P-388 Murine Leukemia Cells. Orient J Chem 2019;35(1). Available from: https://bit.ly/2ToSuzg |
Introduction
Meliaceae family is a woody plant that grows in the tropics consisting of 51 genera and approximately 550 species. Meliaceae plant has been reported to contain many interesting biologically active compounds related to agriculture and health, such as insecticidal, antifeedant, plant growth regulator, mosquito repellent, larvicidal, antimalarial, antiviral, antioxidant, anticancer, antibacterial, antifungal and inflammatory.1-3 The largest genus in the Meliaceae family is Aglaia, which is about 130 species. Aglaia is a tropical plant that distributed throughout Western Pacific, Indonesia, Malaysia and India.4,5 The Aglaia species are commonly found in tropical rainforests in Southeast Asia (Malaysia and Indonesia) with a dozen or more different species living together.4 Phytochemical investigation of plants belonging to Aglaia genus have indicated the presence of sesquiterpenoid,6,7 diterpenoid,8,9 rocaglate derivative,1,10,11 lignan,12,13 dammarane-type triterpenoids,14-18 protolimonoid19 and cycloartane-type triterpenoids.16,20-22 One of the Aglaia plants that only grown in Indonesia is A. glabrata which indigenous to East Kalimantan Province, Indonesia. Presently, isolation, structure determination and cytotoxic activity of triterpenoids 1-4 from A. glabrata are reported.
Materials and Methods
General Experimental Procedure
The melting point values of triterpenoids were obtained using a electrothermal apparatus with uncorrected measurement. A Perkin-Elmer 1760X spectrophotometer was used to obtained infrared spectra of compounds in pellets of potassium bromide. HRTOFMS spectra were recorded on Water, Qtof HR-MS XEVotm mass spectrometer. All NMR spectra are measured on JEOL NMR A-500 MHz with an internal standard of tetramethylsilane. All chromatography techniques, including vacuum liquid chromatography (VLC) and column chromatography (CC), were undertaken on silica gel (SiO2, Merck, Darmstadt, Germany). TLC analysis was done on Kieselgel 60 F254 (Merck) plates; visualization was performed with UV light and plates were sprayed with 10% H2SO4 solution in ethanol and heated.
Plant Material
Dried bark of A. glabrata were obtained in August 2016 from Bogor Botanical Garden, Bogor, West Java, Indonesia. A voucher specimen was deposited at the Bogoriense Herbarium (code Bo-1288718).
Cytotoxic Activity
Determination of cytotoxic activities was undertaken based on the MTT assay explained in the previous reports.18,19 The P-388 murine leukemia cells were seeded into 96-well plates at an initial cell density of approximately 3 x 104 cells cm-3. After 24 hours of incubation for cell attachment and growth, varying concentrations of samples were added. The compounds were first dissolved in DMSO (dimethyl sulfoxide) at the required concentration. Six concentrations were prepared using PBS (phosphoric buffer solution, pH = 7.30 – 7.65). Control wells received only DMSO. The assay was terminated after 48 hours incubation period by adding MTT reagent [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] and the incubation was continued for another 4 hours, in which the MTT-stop solution containing SDS (sodium dodecyl sulphate) was added and another 24 hours incubation was conducted. An optical density was read by using a micro plate reader at 550 nm. IC50 values were taken from the plotted graph of percentage live cells compared to control (%), receiving only PBS and DMSO, versus the tested concentration of compounds (µg/mL). The IC50 value is the concentration required for 50% growth inhibition. Each assay and analysis was run in triplicate and averaged.
Results
Extraction and Isolation
Dried powder of A. glabrata (2.5 kg) were extracted by maceration with 80% aqueous methanol. The extract was partitioned with respective n-hexane and EtOAc. Fraction C of n-hexane extract was VLC-chromatographed twice to yield dammardienon (1, 13.6 mg).23 The second fraction (fraction G) was VLC-chromatographed two times, followed by crystallization to result in aglaiabbreviatin E (2, 12.4 mg).17
Two fractions of EtOAc extract (Fraction J and N) were chromatographed using successive VLC techniques and were followed recrystallization, yielding respective dammar-20,25-dien-3b,24-diol (3, 23.4 mg)24 and dammar-24-en-3β,20-diol (4, 12.4 mg).25,26 The structural determination of all compounds was undertaken on the basis of spectroscopic methods (UV, IR, MS, 1D and 2D NMR) and comparison with those reported in the literature. All of these data can be seen in Table 1 and 2.
Table 1: Characterization data for compounds 1-4.
| Compound | Physical state | Melting point (oC) | m/z [M+H]+ | IR in KBrnmax (cm-1) |
| 1 | Colorless needle crystals | 141-143 | 425.3698 (calcd. for C30H48O, m/z 424.3695) | 3082, 2949, 1705, 1641, 1460 |
| 2 | Whiteness amorphous powder | – | 441.3872 (calcd. for C30H49O2, m/z 441.3878) | 3431, 2943, 2870, 1725, 1642, 1380, 1245 |
| 3 | Colorless needle crystals | 167-169 | 443.3811 (calcd. for C30H48O, m/z 442.3818) | 3430, 2949, 1460, 1380, 1050 |
| 4 | White solid | – | 445.3970 (calcd. for C30H52O2 m/z 444.3977) | 3369, 2939, 1639, 1458, 1109 |
Discussion
The barks of A. glabrata were macerated with aqueous MeOH at room temperature and the macerated residue was fractionated with respective n-hexane, EtOAc and n-BuOH. Repetitive SiO2 CC. of the n-hexane and EtOAc fractions afforded four dammarane-type triterpenoids 1-4 (Figure 1).
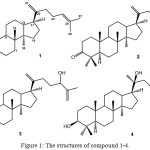 |
Figure 1: The structures of compound 1-4. |
MS data of compound 1 showed that there was a hydrogen deficiency index of seven. The infrared spectra displayed absorbance bands for an aliphatic moiety (3082 and 2949 cm-1), a ketonic carbonyl (1705 cm-1) and an olefin (1645 cm-1). The proton NMR analysis displayed seven methyl protons at δH 0.87, 0.95, 1.00, 1.05, 1.08, 1.61 and 1.68, which corresponding to tertiary Me-18, Me-19, Me-26, Me-27, Me-28, Me-29, respectively. There are signals of a sp2 methine proton at chemical shift of δH 5.11 and sp2 methylene protons at chemical shifts of δH 4.71 and 4.75, found in the spectrum. Thirty carbon signals were read in the carbon NMR spectra after associated with DEPT analysis, which consisted of seven tertiary methyls, ten methylenes of sp3, four methines of sp3, four quaternary carbons of sp3, one carbonyl (δC 217.6), one sp2 methylene (δC 106.9) one sp2 methines (δC 125.1) and two sp2 quaternary carbons (δC 132.0 and 151.4). Four remaining of hydrogen deficiency index was corresponding to four ring dammarane-type triterpenoid.18,19 The presence of dammarane-type in 1 was also supported by a combined experiment of 1H–1H COSY and HMBC (Figure 2). Correlations of an olefinic proton signals at δH 5.11 (H-24) to δC 132.0 (C-25) and δC 28.0 (C-23) and at δH 1.68 (Me-26) and δH 1.61 (Me-27) to δC 132.0 (C-25) strongly supported that double bond was positioned at C-24/C-25. A double bond at terminal C-20/C-2120,21 was deduced from correlations between two protons at δH 4.71 and 4.75 and C-20 (δC 151.4), δC 35.1 (C-22) and δC 48.1 (C-17). Correlations of Me-28 (δH 1.05), 1.92 (H-2a) and 2.40 (H-2b) to carbonyl signal at δC 217.6, indicated the position of carbonyl ketone at C-3. NMR data of 1 were superimposed with those of dammardienon,23 consequently, compound 1 was determined as a dammardienon.
Similar to compound 1, there was a hydrogen deficiency index of seven in compound 2, based on the MS data. The infrared spectrum displayed absorbance band for groups of an alcohol (3431 cm-1), an aliphatic (2943 and 2870 cm-1), a ketonic carbonyl (1725 cm-1), olefine (1642 cm-1) and ether moieties (1245 cm-1). The molecular weight of 2 has 17 atomic mass unit higher than 1, describing an additional OH group in 2. The NMR spectra of 2 have similarity with spectra of 1.
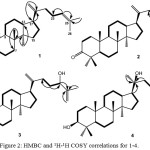 |
Figure 2: HMBC and 1H-1H COSY correlations for 1-4. |
The main differences are the presence of newly double bond at δH 5.64 and 5.70 and one signal of oxygenated carbon at δC 70.7 instead of a proton signal of olefin at δH 5.11, δC 125.1, showing a double bond in new position and a hydroxyl group in 2. HMBC correlations of olefinic protons at δH 5.64 to δC 37.2 (C-22) and δC125.6 (C-23) and δH 5.70 to δC 125.6 (C-23) and δC 70.7 (C-25) were used to determine the newly double bond.23,24 Additionally, correlations from two methyls at Me-26 (δC 28.4) and Me-27 (δC 28.4) to an oxygenated carbon (δC 70.7) were used to determine the newly hydroxyl group at C-25. NMR data of 2 were superimposed with those of aglaiabbreviatin E,17 which strongly recommended that compound 2 was aglaiabbreviatin E.
A molecular formula of compound 3 suggested a hydrogen deficiency index of six, based on its MS data. The IR spectrum showed functional group wave numbers at 3430 cm-1 (hydroxyl), 2949 and 2875 cm-1 (aliphatic), 1610 cm-1 (olefinic), 1450 and 1354 cm-1 (gem-dimethyl) and 1050 cm-1 (ether). The NMR spectral data of 3 were superimposed with spectral data of dammaradienon (1), unless the absence of a ketonic carbonyl (δC 218.4) at C-3 and the presence of an oxymethine (δC 76.5), which were confirmed by HMBC analysis through correlations of CH2-2 (δH 1.86 and 2.01) and two methyl signals (δH 1.05) to C-3 (δC 76.5). Another difference is the presence of an oxygenated sp3 methines at δH 4.20 and δC 70.5. A proton signal of oxygenated methine at δH 4.20 was correlated to C-23 (δC 25.9) and quaternary sp2 carbon at C-25 (δC 141.6), suggesting that a secondary hydroxyl group was located at C-24. Another double bond was assigned to be at position C-25/C-26 based on the correlation of terminal double bond at δH 5.10 and 5.20 as well as a methyl at C-27 (δH 1.70) to sp2 quaternary carbon at C-25 (δC 141.6). NMR data of 3 were highly overlapped to those of dammar-20,25-dien-3b,24-diol,24 eventually, compound 3 was identified as a dammar-20,25-dien-3b,24-diol.
A hydrogen deficiency index of five was also found for compound 4 based on the MS data. IR wave numbers of 3369 cm-1, 2939 cm-1, 1639 cm-1, 1458 and 1230 cm-1, and 1109 cm-1 were analyzed for respective hydroxyl, aliphatic, olefinic, gem-dimethyl, and ether groups. NMR spectral data of 4 were similar to spectral data of dammar-20,25-dien-3b,24-diol (3), except the absence of one of two terminal double bonds at δH 4.85, 4.90 and one oxygenated methine at δH 4.20 and appearance of newly an olefinic double bond at δH 5.10 and δC 124.8 and 131.7 and a tertiary methyl at δH 1.13, δC 25.5, suggesting that compound 4 was the lack of one of the olefinic moiety and appearance of a hydroxyl group. In the HMBC spectrum, correlation from tertiary methyl at δH 1.13 and sp3 methine at δH 1.69 to sp3 oxygenated carbon at δC 75.5, were used to determine the C-20 position of the tertiary hydroxy group. A remaining olefine was positioned at C-24/C-25 that was determined based on the correlation of an olefinic proton at δH 5.10 and tertiary methyl at δH 1.66 to sp2 tertiary carbon at C-25 (δC 131.7). The NMR spectra data of 4 were superimposed with those of dammar-24-en-3β,20-diol,25,26 therefore, compound 4 was characterized as a dammar-24-en-3β,20-diol.
The cytotoxic properties of compounds 1-4 were tested towards P-388 murine leukemia cells using a modified method priory described.18,20,26 Artonin E was used as a positive control with IC50 0.68 µM.27 The results of cytotoxic assay of isolated compounds 1-4 (Table 3), showed that dammar-24-en-3b, 20-diol (4), having two hydroxy groups and one double bond, has the strongest activity among all, whereas dammardienon (1) has the weakest activity, suggesting that the presence of hydroxyl and double bond moieties are significant structural element for the cytotoxic activity.
Tabel 2: 1H (CDCl3, 600 MHz) and 13C NMR (CDCl3, 150 MHz) data for Compounds 1-4.
| Position | 1 | 2 | 3 | 4 | |||||
| δC (mult.) | δH, mult., (J Hz) | δC (mult.) | δH, mult.,(J Hz) | δC(mult.) | δH, mult.,(J Hz) | δC(mult.) | δH, mult., (J Hz) | ||
| 1a | 40.5 (t) | 1.44, dd (2.8, 7.2) | 40.1 (t) | 1.44, dd (1.7, 9.4) | 37.9 (t) | 1.40, dd (1.9, 7.5) | 37.2 (t) | 1.37, ddd (1.6, 2.8, 8.6) | |
| 1b | 1.93, ddd (2.4, 6.2, 7.2) | 1.90, ddd (1.4, 4.8, 9.4) | 1.44, ddd (2.1, 4.7, 7.5) | 1.54, ddd (1.2, 4.5, 8.6) | |||||
| 2a | 35.1 (t) | 1.92, dd (3.1, 8.2) | 35.2 (t) | 2.45, dd (3.6, 14.0) | 33.6 (t) | 2.01, dd (2.1, 8.4) | 34.2 (t) | 1.63, ddd (1.6, 4.5, 9.8) | |
| 2b | 2.40, ddd (2.7, 5.6, 8.2) | 2.70 ,dt (14.0, 5.2) | 1.86, ddd (1.7, 6.4, 8.4) | 1.65, ddd (1.2, 4.5, 9.8) | |||||
| 3 | 217.6 (s) | 217.8 (s) | 76.5 (d) | 3.39, t (2.6) | 77.6 (d) | 3.37, dd (1.6, 4.5) | |||
| 4 | 46.8 (s) | 46.9 (s) | 49.7 (s) | 48.1 (s) | |||||
| 5 | 54.7 (d) | 1.39 ,dd (2.9, 4.5) | 55.2 (d) | 1.36, dd (2.3, 7.6) | 55.2 (d) | 1.26, dd (2.6, 4.8) | 54.9 (s) | 1.43, t (7.2) | |
| 6a | 20.1 (t) | 1.46, dd (4.5, 7.4) | 19.4 (t) | 1.45, ddd (2.3, 7.6, 10.6) | 18.6 (t) | 1.41, ddd (2.6, 6.4, 9.5) | 18.3 (t) | 1.58, dd (7.2, 9.8) | |
| 6b | 1.70, ddd (2.9, 5.1, 7.4) | 1.54, ddd (1.9, 7.6, 10.6) | 1.61, ddd (1.9, 5.7, 9.5) | 1.48, ddd (2.1, 7.2, 9.8) | |||||
| 7a | 35.2 (t) | 1.33, dd (5.1, 8.4) | 34.8 (t) | 1.68 dd (1.8, 9.8) | 35.3 (t) | 1.29, ddd (1.9, 5.7, 10.4) | 35.2 (t) | 1.44, ddd (2.1, 7.2, 11.2) | |
| 7b | 1.58, ddd (2.8, 4.5, 8.4) | 1.89, ddd (1.5, 2.5, 9.8) | 1.54, ddd (1.8, 6.8, 10.4) | 1.75, ddd (2.1, 5.4, 11.2) | |||||
| 8 | 30.9 (s) | 30.4 (s) | 30.6 (s) | 30.7 (s) | |||||
| 9 | 40.6 (d) | 1.41, t (5.3) | 40.3 (d) | 1.40,dd (2.3, 5.7) | 38.9 (d) | 1.45, t (6.8) | 40.3 (d) | 1.72, t (6.4) | |
| 10 | 50.5 (s) | 50.3 (s) | 50.6 (s) | 50.4 (s) | |||||
| 11a | 21.9 (t) | 1.54, dd (5.3, 8.2) | 21.8 (t) | 1.40, dd (5.7, 11.6) | 22.3 (t) | 1.54, ddd (1.8, 6.8, 8.6) | 21.4 (t) | 1.82, dd (6.4, 8.8) | |
| 11b | 1.83, ddd (2.1, 5.3, 8.2) | 1.50, ddd (2.3, 5.7, 11.6) | 1.62, ddd (1.6, 4.7, 8.6) | 1.96, ddd (1.7, 6.4, 8.8) | |||||
| 12a | 28.8 (t) | 1.45, dd (3.6, 7.8) | 28.8 (t) | 1.07, dd (2.3, 9.8) | 27.7 (t) | 1.15, ddd (1.8, 4.7, 10.8) | 27.4 (t) | 1.83, ddd (1.7, 6.4, 12.2) | |
| 12b | 1.91, ddd (2.1, 3.6, 7.8) | 1.58, ddd (2.3, 6.2, 9.8) | 1.20, ddd (1.6, 4.7, 10.8) | 2.01, ddd (1.4, 5.6, 12.2) | |||||
| 13 | 46.1 (d) | 1.69, dd (7.8, 8.1) | 47.5 (d) | 2.24, dd (1.8, 8.4) | 42.8 (d) | 1.61, t (7.2) | 42.3 (d) | 1.78, t (4.6) | |
| 14 | 50.2 (s) | 49.8 (s) | 49.7 (s) | 50.4 (s) | |||||
| 15a | 30.9 (t) | 1.61, dd (1.8, 8.5) | 31.1 (t) | 1.58, dd (1.8, 9.5) | 28.5 (t) | 1.45, ddd (1.5, 3.4, 9.6) | 31.2 (t) | 2.04, dd (7.2, 11.4) | |
| 15b | 2.01, ddd (1.8, 5.6, 7.8) | 1.60, ddd (1.8, 4.8, 9.5) | 1.06, ddd (1.8, 3.4, 9.6) | 2.45, ddd (2.4, 7.2, 11.4) | |||||
| 16a | 26.0 (t) | 1.51, ddd (1.9, 2.7, 7.8) | 25.9 (t) | 1.40, dd (1.6, 10.4) | 22.7 (t) | 1.88, ddd (3.4, 6.5, 11.2) | 22.6 (t) | 2.77, ddd (2.4, 7.2, 10.8) | |
| 16b | 1.98, ddd (2.7, 7.8, 8.1) | 1.90, ddd (1.6, 2.5, 10.4) | 1.92, ddd (1.8, 3.4, 11.2) | 2.32, ddd (2.4, 4.2, 10.8) | |||||
| 17 | 48.1 (d) | 2.20, ddd (2.7, 4.5, 7.8) | 46.6 (d) | 1.68, dd (3.2, 10.4) | 49.9 (d) | 1.47, dd (1.8, 3.4) | 49.8 (d) | 2.29, dd (2.4, 7.2) | |
| 18 | 16.0 (q) | 1.00, s | 15.7 (q) | 1.01, s | 16.1 (q) | 0.96, s | 16.6 (q) | 0.93, s | |
| 19 | 17.1 (q) | 0.95, s | 16.8 (q) | 1.08, s | 16.7 (q) | 0.95, s | 16.1 (q) | 0.90, s | |
| C-20 | 151.3 (s) | 124.7 (s) | 75.5 (s) | ||||||
| 21a | 106.9 (t) | 4.71, d (7.8) | 109.0 (t) | 4.88, s | 140.1 (t) | 4.85, d (5.6) | 25.5 (q) | 1.03, s | |
| 21b | 4.75, d (7.8) | 4.95, s | 4.90, d (5.6) | ||||||
| 22a | 35.1 (t) | 2.04,dd (2.4, 6.7) | 37.2 (t) | 2.66, d (4.2) | 35.7 (t) | 2.66, d (3.8) | 40.6 (t) | 1.34, dd (2.1, 8.4) | |
| 22b | 2.32, dd (1.8, 6.7) | 2.70, d (4.2) | 2.70, d (3.8) | 1.72, ddd (2.1, 3.6, 8.4) | |||||
| 23 a | 28.0 (t) | 1.96, dd (1.5, 5.8) | 125.6 (d) | 5.64, dd (6.5, 8.2) | 25.9 (t) | 2.80, m | 22.6 (t) | 1.85, dd (2.1, 9.7) | |
| 23 b | 2.98, dd (2.1, 4.6) | 2.01, ddd (1.7, 5.4, 9.7) | |||||||
| 24 | 125.1 (d) | 5.11, dd (2.2, 5.8) | 139.0 (d) | 5.70, dd (6.5,8.2) | 70.5 (d) | 4.20, dd (2.1, 4.6) | 125.2 (d) | 5.10, dd (1.7, 5.4) | |
| 25 | 132.0 (s) | 70.7 (s) | 141.6 (s) | 132.1 (s) | |||||
| 26 | 25.9 (q) | 1.68, s | 28.4 (q) | 1.32, s | 124.2 (t) | 5.10, d (4.6)5.20, d (4.6) | 25.9 (q) | 1.86, s | |
| 27 | 17.9 (q) | 1.61, s | 28.4 (q) | 1.32, s | 17.6 (q) | 1.70, s | 17.8 (q) | 1.78, s | |
| 28 | 20.9 (q) | 1.05, s | 25.7 (q) | 0.94, s | 22.1 (q) | 1.05, s | 28.4 (q) | 0.94, s | |
| 29 | 27.1 (q) | 1.08, s | 21.5 (q) | 1.04, s | 26.0 (q) | 1.05, s | 23.1 (q) | 0.88, s | |
| 30 | 15.9 (q) | 0.87, s | 16.0 (q) | 0.87, s | 16.1 (q) | 1.02, s | 17.2 (q) | 0.96, s | |
Table 3: Cytotoxicity activity of compounds 1–4.
| Compounds | IC50 (mg/mL) |
| Dammardienon (1)Aglaiabbreviatin E (2) | 40.25 ± 0.1234.25 ± 0.12 |
| Dammar-20,25-dien-3b,24-diol (3) | 32.20 ± 0.10 |
| Dammar-24-en-3b,20-diol (4) | 9.45 ± 0.10 |
Conclusion
Four dammarane-type triterpenoids, 1-4, were isolated from the bark of A. glabrata, for the first time in this species. These findings support the presence of dammarane triterpenoid in the Aglaia genus. In dammarane-type triterpenoids, hydroxyl and double bond groups are important requirements for the cytotoxic properties.
Acknowledgments
This investigation was financially supported by Directorate General of Scientific Resources, Technology and Higher Education, Ministry of Research, Technology and Higher Education, Indonesia (World Class Professor Grant, No 123.38/D2.3/KP/2018 by Unang Supratman).
References
- Nugroho, B.W.; Edrada, R.A.; Wray, V.; Witte, L.; Bringmann, G.; Gehling, M.; Proksch, P. Phytochemistry. 1999, 51, 367-376.
- Omar, S.; Zhang, J.; Kinnon, M.S.; Leaman, D.; Arnason, J.T.; Philogen, B.J.R. Current Top Medicinal Chemistry. 2003, 3(2), 133-139.
- Nakatani, M.; Abdelgalell, S.A.M.; Sand, M.M.G.; Huang, R.C.; Doe, N.; Iwagawa, T. Phytochemistry. 2004, 65, 2833-2841.
- Pannell, C.M. Taxonomic Monograph of the Genus Aglaia Lour. (Meliaceae). Kew Bulletin Additional Series XVI; HMSO: Kew, Richmond, Surrey, UK. 1992, pp, 128-150.
- Inada, A.; Sorano, T.; Murata, H.; Inatomi, Y.; Darnaedi, D.; Nakanishi, T. Chem. Pharm. Bull. 2001, 49(9), 1226-1228.
- Joycharat, N.; Plodpai, P.; Panthong, K.; Yingyongnarongkul, B.; Voravuthikunchai, S.P. Can. J. Chem. 2010, 88, 937–944.
-
Liu, S.; Liu, S.B.; Zuo, W.; Guo, Z.; Mei, W.; Dai, H. Fitoterapia. 2014, 92, 93–99.
- Cai, X., Wang, Y., Zhao, P., Li, Y., Luo, X. Dolabellane diterpenoids from Aglaia odorata. Phytochemistry. 2010, 71, 1020–1024.
- Yodsaoue, O.; Sonprasit, J.; Karalai, C.; Ponglimanont, C.; Tewtrakul, S.; Chantrapromma, S. Phytochemistry. 2012, 76, 83-91.
- Ishibashi, F.C.; Satasook, M.B.; Towers, G.H.N. Phytochemistry. 1993, 32, 307-310.
- Wu, T.S.; Liou, M.J.; Kuoh, C.S.; Teng, C.M.; Nagao, T.; Lee, K.H. J. Nat. Prod. 1997, 60, 606-608.
- Wang, B.; Ebel, R.; Wang, C.; Edrada, R.U.; Wray V.; Proksch, P. J. Nat. Prod. 2004, 67, 682-684..
- Sianturi, J.; Purnamasari, M.; Darwati.; Harneti, D.; Mayanti, T.; Supratman, U.; Awang, K.; Hayashi, H. Phytochemistry Letters. 2015, 13, 297–301.
- Roux, D.T.; Martin, T.; Adeline, T.; Sevenet, H.; Hadi.; Pais, M. Phytochemistry. 1998, 49(6), 1745-1748.
- Khalit, M.; Martin, M.T.; Leroy, E.; Tempete, C.; Sevenet, T.; Awang, K.; Pais, M. Phytochemistry. 1999, 51, 1031-1037.
- Xie, B.J.; Yang, S.P.; Chen, H.D.; Yue, J.M. J. Nat. Prod. 2007, 70, 1532-1535.
- Zhang, F.; Wang, J.S.; Gu, Y.C.; Kong, L.Y. J. Nat. Prod. 2010, 73, 2042-2046.
- Harneti, D.; Tjokronegoro, R.; Safari, A.; Supratman, U.; Loong, X.M.; Mukhtar, M,R.; Mohamad, K.; Awang, K.; Hayashi, H. Phytochemistry Letters. 2012, 5, 496–499.
- Farabi, K.; Harneti, D.; Nurlelasari.; Maharani, R.; Hidayat, A.C.; Awang, K.; Supratman, U.; Shiono, Y. Phytochemistry Letters. 2017, 21. 211-215.
- Awang, K.; Loong, X.M.; Leong, K.H.; Supratman, U.; Litaudon, M.; Mukhtar, M.R.; Mohamad, K. Fitoterapia. 2012, 83, 1391-1395.
- Leong, K.H.; Looi, C.Y.; Loong, X.M.; Cheah, F.K.; Supratman, U.; Litaudon, M.; Mustafa, M.R.; Awang, K. PLOS ONE. 2016, 4, 1-17.
- Teles, Y.C.F.; Gomes, R.A.; Olivera, M.S.; de Lucena, K.L.; do Nascimento, J.S.; Agra, M.F.; Igoli, J.O.; Gray, A.L.; de Souza, M.F.V. Quin Vova. 2014, 37(9), 1491-1495.
- Grande, M.; Torres, P.; Piera, F.; Bellido, I. Phytochemistry, 1992, 31(5), 1826-1828.
- Phongmaykin, J.; Kumamoto, T.; Ishikawa, T.; Suttisri, R.; Saifah, E. Arch. Pharmacetical Research. 2008, 31, 21-27.
- Hidayat, A.T.; Farabi, K.; Harneti, D.; Nurlelasari.; Maharani, R.; Nurfarida, I.; Supratman, U.; Shiono, Y. Indones. J. Chem. 2018, 18(1), 35–42.
- Alley, M.C.; Scudiero, D.A.; Monks, A.; Hursey, M.L.; Czerwinski, M.J.; Fine, D.L.; Abbott, B.J.; Mayo, J.G.; Shoemaker, R.H.; Boyd, M.R. Cancer Resesarch. 1988, 48, 589–601.
- Hakim, E.H.; Achmad, S.A.; Juliawaty, L.D.; Makmur, L.; Syah, Y.M.; Aimi, A.; Kitajima, M.; Takayama, H.; Ghisalberti, E.L. J. Nat. Med. 2007, 61(2), 229–236.

This work is licensed under a Creative Commons Attribution 4.0 International License.









