The Performance of P-Aminosalicylic Acid As Reducing and Stabilizing Agent in Silver Nanoparticles Synthesis
Dian Susanthy, Sri Juari Santosa* and Eko Sri Kunarti
and Eko Sri Kunarti
Department of Chemistry, Faculty of Mathematics and Natural Sciences, Universitas Gadjah Mada, Sekip Utara Bulaksumur, Yogyakarta, 55281, Indonesia.
Corresponding Author E-mail: sjuari@ugm.ac.id
DOI : http://dx.doi.org/10.13005/ojc/350106
Article Received on : 06-10-2018
Article Accepted on : 07-01-2019
Article Published : 11 Feb 2019
In this study, silver nanoparticles (AgNPs) were successfully synthesized using p-aminosalicylic acid as a reducing and stabilizing agent simultaneously. The AgNPs was synthesized by mixing silver nitrate solution as a precursor with the pH adjusted by p-aminosalicylic acid solution and heating it in a boiling water bath. The formed AgNPs were analyzed using UV-Vis spectrophotometry to evaluate their SPR absorbance in the wavelength range of 400-500 nm. The optimum reaction time is 10 min and the optimum pH is 11. The AgNPs with the optimum synthesis condition have average size of 32.3 nm when characterized using PSA, spherical morphology when characterized using TEM, and face-centered cubic crystal when characterized using XRD. The formed AgNPs had good stability for more than 2 months. The mechanism of silver ion reduction and AgNPs stabilization by p-aminosalicylic acid were also proposed in the paper based on the FTIR analysis result.
KEYWORDS:P-Aminosalicylic Acid; Reducing Agent; Silver Nanoparticles; Stabilizing Agent
Download this article as:| Copy the following to cite this article: Susanthy D, Santosa S. J, Kunarti E. K. The Performance of P-Aminosalicylic Acid As Reducing and Stabilizing Agent in Silver Nanoparticles Synthesis. Orient J Chem 2019;35(1). |
| Copy the following to cite this URL: Susanthy D, Santosa S. J, Kunarti E. K. The Performance of P-Aminosalicylic Acid As Reducing and Stabilizing Agent in Silver Nanoparticles Synthesis. Orient J Chem 2019;35(1). Available from: https://bit.ly/2SGQrt3 |
Introduction
Silver nanoparticles (AgNPs) are very popular nanomaterial. In addition to the abundance of silver in the earth crust, silver nanoparticles also have a broad spectrum of bactericidal activity, surface plasmon resonance absorption, and low cost of manufacturing.1 For these reasons, AgNPs have been used in many application, such as antimicrobial agent,2,3,4 part of environmental treatments, such as air, water, and surface disinfection,5 and as chemical sensors for ammonia,6 chlorine dioxide,7 herbicides,8 copper ions,9 and lead.10
In this study, the AgNPs synthesized by chemical reduction method. Chemical reduction method is preferable because of its convenience factor, relatively low cost, and likely to be produced on a large scale.11 The reducing agent will reduce the positive silver ion from the precursor solution to become zero charged silver particles which further will become AgNPs. The stabilizing agent was used to form a system, which can protect the AgNPs and prevent aggregation.12 Many studies have been conducted by using different chemicals as reducing and stabilizing agents.13,14,15 However, AgNPs synthesis by using only one chemical as both reducing and stabilizing agent is preferable than using two different chemicals. Some previous study had been conducted by using benzoic acid derivatives with hydroxy phenolic group as reducing and stabilizing agent in the AgNPs synthesis.16,17,18 However, the use of benzoic acid derivatives with amino and hydroxyl phenolic groups has not been performed yet.
In order to discover the role of each functional group in benzoic acid derivative, p-aminosalicylic acid was used in this research as reducing and stabilizing agent in AgNPs synthesis. The amino and hydroxyl phenolic groups were predicted to reduce the Ag+ ions to become Ag0, and the carboxylate groups will stabilize the formed AgNPs. This study critically examines the effect of the amino group addition or substitution on the benzoic acid derivatives as both reducing agents of Ag+ to Ag0 and as stabilizing agents for the formed AgNPs. Furthermore, a comparison was made with existing research which used other benzoic acid derivative in AgNPs syntheses, such as hydroxybenzoic acid and dihydroxibenzoic acid. The parameter examined includes the particle size, shape, and stability. In addition, the AgNPs formation and stability mechanism by p-aminosalicylic acid were also proposed based on the FTIR spectra of synthesized AgNPs.
Experimental
Materials
All reagents were used as received without further purification. Silver nitrate (Merck) as precursor of AgNPs, p-aminosalicylic acid (Sigma Aldrich) as reducing and stabilizing agent, nitric acid and sodium hydroxide (Merck) as pH adjuster.
Synthesis of AgNPs
Silver nitrate solution (0.3 × 10-3 mol/L) was added to pH-adjusted p-aminosalicylic acid (10 × 10-3 mol/L) in a test tube with volume ratio of 1:1 and homogenized. The mixture was then heated in a boiling water bath for several minutes until it turned yellow which indicates that the AgNPs were formed. After the reaction was finished, the mixture was cooled in tap water and transferred to a small bottle for further analysis. For reaction optimization, some reaction parameters were varied, such as pH (5-13), reaction time (0.5, 10, 15, 20, 25, 30 min), and mole ratio of the reactants.
Characterization of AgNPs
Synthesized AgNPs were characterized using UV-Vis spectrophotometer (Shimadzu UV-Pharma Spec 1700) in order to measure their SPR absorbance. The scanning was performed at 300-800 nm wavelength range and quartz cuvette with 1 cm optical path length. The scanning was performed in fast speed with interval of 1 nm.
For TEM analysis (JEOL JEM-1400), the AgNPs colloid was immersed by copper grid. The immersed copper grid was then dried at room temperature. The image was taken by using 120 kV accelerating voltage. The TEM image was further analyzed using imageJ software in order to observe the particle size and measure the particle size of AgNPs.
AgNPs was also characterized by X-Ray Diffraction (Rigaku) to confirm the crystal form of AgNPs. The XRD spectra was also used to calculate the particle size of the AgNPs by using Debye-Scherrer Equation (Equation 1) where d is average particle size (nm), K is constant (0.9), λ is the wavelength of X-ray (nm), FWHM is the full width at maxima of the peak, and θ is the Bragg angle (°).19
![]()
Particle Size Analysis (Horiba SZ-100) also performed for the synthesized AgNPs to measure the particle size of AgNPs and their polydispersity index using dynamic light scattering method. The temperature of the holder is 25°C with scattering angle of 90. Furthermore, the zeta potential measurement also performed using the same instrument in order to evaluate the AgNPs stability.
The FTIR analysis (Shimadzu FTIR Prestige-21) was also performed in order to predict the reduction mechanism in the AgNPs synthesis. The FTIR analysis was performed for synthesized AgNPs and p-aminosalicylic acid. The AgNPs was separated from their solution using centrifuge with speed of 13000 rpm. The separated AgNPs were then dried in the oven at temperature of 65°C. The dried AgNPs were then mixed with KBr and formed into pellet. The pellet was analyzed in wavelength range of 400-4000 cm-1.
Results and Discussion
The Effect of Acidity in AgNPs Synthesis
Solution acidity is one important parameter for the synthesis of AgNPs. In this study, the pH of p-aminosalicylic acid was varied from 5 to 13. The solution could not be more acidic because p-aminosalicylic acid could not be dissolved when pH was lower than 5. It could not be more basic since the silver ions in the silver nitrate solution as precursor would be reduced to solid silver above pH 13. The formation of AgNPs was indicated by the occurrence of absorbance band in the wavelength range of 400-500 nm. This band was known as SPR absorbance of AgNPs.20
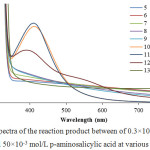 |
Figure 1: The spectra of the reaction product between of 0.3×10-3 mol/L AgNO3 and 50×10-3 mol/L p-aminosalicylic acid at various pH. |
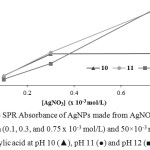 |
Figure 2: The SPR Absorbance of AgNPs made from AgNO3 with various concentration (0.1, 0.3, and 0.75 x 10-3 mol/L) and 50×10-3 mol/L p-aminosalicylic acid at pH 10 (▲), pH 11 (●) and pH 12 (■). |
Based on the spectra in Figure 1, it can be shown that the reaction can occur at pH of 10, 11, and 12. At pH 10, the absorption band was at 411 nm with absorbance of 1.548. At pH 11, the absorbance band was also at 411 nm with an absorbance of 1.614 while at pH 12 the absorbance band was at 392 nm with absorbance of 1.069. In order to make sure the optimum pH condition, the concentration of silver nitrate solution was varied (0.1, 0.3, and 0.75 x 10-3 mol/L) for each pH in order to evaluate the reducing performance and stabilizing performance of p-aminosalicylic acid in each pH. The result is shown in Figure 2. It can be seen that in pH 11, the formed silver nanoparticles in almost every precursor concentration gave the highest absorbance value than another pH. It means that p-aminosalicylic acid has the best reducing performance in pH 11.
Furthermore, the result of stability evaluation showed that the silver nanoparticle which synthesized in pH 11 has the better stability than nanoparticles which synthesized in pH 12 because silver nanoparticles which synthesized in pH 12 settled down earlier than silver nanoparticles which synthesized in pH 11. Increasing the pH value of p-aminosalicylic acid can improve its reducing performance, but decrease its stabilizing performance on the formed silver nanoparticles. Vice versa, decreasing the pH value of p-aminosalicylic acid can improve its stabilizing performance on formed silver nanoparticles, but decrease its reducing performance. Based on these reasons, it can be concluded that pH 11 is the optimum pH for the silver nanoparticles synthesis with p-aminosalicylic acid as reducing and stabilizing agent.
The same optimum pH was also obtained for several reducing agents, which are the derivatives of the benzoic acid, such as hydroxybenzoic acid,16,17 dihydroxibenzoic acid,18 and aminobenzoic acid.21 The synthesis of AgNPs by using derivatives of benzoic acid are usually conducted under basic conditions. It happens because the benzoic acid group will be deprotonated and the formed negative charge can interact easily with the positive metal ion.22 Under the basic condition, amino and hydroxyphenolic groups become more negative because their proton atoms are attracted by the base. This condition also will make amino and hydroxyphenolic groups able to interact more easily with the positive metal ion.
The Effect of Reaction Time in AgNPs Synthesis
For the time optimization, the reduction reaction was monitored at the regular time interval using UV-Vis spectrophotometry from 0 to 45 min. The result is shown in Figure 3. It can be seen that the optimum reaction time is 10 min since the increase in reaction time above 10 min did not give a significant increase in the SPR absorbance.
Compared to silver nanoparticles synthesis with using o-hydroxybenzoic acid as reducing agent which took about 2 hours to obtain silver nanoparticle SPR absorbance around 0.4 with the same pH, reactant concentration, and reaction temperature16 and AgNPs synthesized by p-aminobenzoic acid which needs 30 minutes to obtain its maximum SPR absorbance around 0.9 with the same pH, silver nitrate concentration, and reaction temperature,21 p-aminosalicylic acid has greater reducing performance since it gave higher SPR absorbance of silver nanoparticles in shorter time reaction. On the other hand, compared to silver nanoparticles synthesis with using o,p-dihydroxibenzoic acid as reducing agent,18 the use of p-aminosalicylic acid as reducing agent gave resemblant absorbance.
Based on these comparisons, it can be concluded that the amino or hydroxyl group addition can increase the reducing performance of benzoic acid derivative compound because there will be more active moiety in the compound. However, compared to hydroxyl group, the amino group substitution did not give any significant difference in its reducing performance.
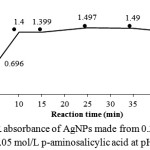 |
Figure 3: SPR absorbance of AgNPs made from 0.3 x 10-3 mol/L AgNO3 and 0.05 mol/L p-aminosalicylic acid at pH 11. |
The Effect of Reactant Mole Ratio in AgNPs Synthesis
The concentration of each reactant is very important in the formation of AgNPs. In this study, varying the reactant mole ratio was done using AgNO3 with concentration of 0.3 x 10-3 mol/L and p-aminosalicylic acid with concentration 0, 2, 4, 6, 8, and 10 x10-3 mol/L. The spectra of the product from the reaction of various mole ratios of AgNO3 to p-aminosalicylic acid are shown in Figure 4. There was 3 highest absorbance value, they are 1.40, 1.42, and 1.41 for mole ratio of 1:20, 1:27, and 1:33 (AgNO3: p-aminosalicylic acid), respectively. It means that p-aminosalicylic acid has reached its maximum reducing performance in mole ratio of 1:27 where 1 molecule of AgNO3 was attracted with 27 molecules of p-aminosa-licylic acid.
The same mole ratio was also used by Susanthy et al. (2017) which used o,p-dihydroxibenzoic acid as reducing and stabilizing agent in AgNPs synthesis.18 But this result was a little bit different with the effective mole ratio of AgNO3 to the p-hydroxyben-zoic acid which has value 1:20.16 It can be concluded that the number of active moiety in reducing agent influence the optimum ratio mole of reactant in AgNPs synthesis.
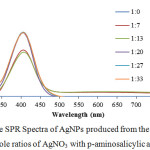 |
Figure 4: The SPR Spectra of AgNPs produced from the reaction of various mole ratios of AgNO3 with p-aminosalicylic acid at pH 11. |
Characterization of AgNPs
TEM
The reducing ability of a reducing agent can be evaluated by the formed particle size. Silver nanoparticles formed through the reduction of Ag2O aggregates which then reduced by reducing agent to become zero charge silver ion and finally become silver nanoparticles.23 Reducing agent with better reducing performance will produce silver nanoparticles with smaller particle size. The TEM image of the optimum AgNPs and its particle size distribution are shown in Figure 5. It can be seen that AgNPs have a spherical shape. The average particle sizes counted from the TEM image are 14.55±7.13 nm.
Compared to the previous study, AgNPs which synthesized using p-aminosalicylic acid is smaller than AgNPs which synthesized using hydroxybenzoic acid16,17 and dihydroxibenzoic acid.18 However, AgNPs in this research are still not uniform, some particles stick to each other because the reduction process had not finished and kept going on even under room condition. Based on these result in the term of the formed particle size, the addition or substitution of amino groups in benzoic acid derivative compound can increase its reducing performance on silver nanoparticle synthesis.
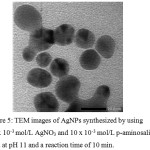 |
Figure 5: TEM images of AgNPs synthesized by using 0.3 x 10-3 mol/L AgNO3 and 10 x 10-3 mol/L p-aminosalicylic acid at pH 11 and a reaction time of 10 min. |
Particle Size Analyzer
The synthesized AgNPs was also analyzed by Particle Size Analyzer to obtain the more accurate particle size. The result showed that the AgNPs which synthesized using p-aminosalycilic acid had average particle size of 32.3 nm with PI value of 0.34. This result was bigger than using TEM because the PSA analyze the whole particle in the solution, including some bigger silver particle which had not become silver nanoparticles, while TEM analyze particular part of the solution. The zeta potential value of the synthesized AgNPs is -21.3 mV. It means that the synthesized AgNPs was capped by negative charge which most likely come from the deprotonized carboxylate moiety. However, the zeta potential value of AgNPs capped by p-aminosalicylic acid was not high enough to be said as stable nanoparticle because it was smaller than 30 mV.24
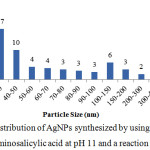 |
Figure 6: Particle size distribution of AgNPs synthesized by using 0.3 x 10-3 mol/L AgNO3 and 10 x 10-3 mol/L p-aminosalicylic acid at pH 11 and a reaction time of 10 min. |
X-Ray Diffraction
The XRD spectra of the synthesized AgNPs was showed in Figure 6. Compared to AgNPs which synthesized with chitosan-polyethylene glycol,25 there were 4 similar peaks, they are peaks in 2θ of 38.10°C, 44.34°C, 64.64°C, and 77.38°C which were characteristics to face-centered cubic of AgNPs.
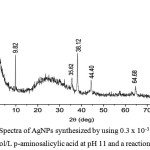 |
Figure 7: XRD Spectra of AgNPs synthesized by using 0.3 x 10-3 mol/L AgNO3 and 10 x 10-3 mol/L p-aminosalicylic acid at pH 11 and a reaction time of 10 min. |
The obtained peaks from XRD spectra also can be used to calculate the particle size of the nanoparticles by using Debye-Scherrer Equation (1) where d is average particle size (nm), K is constant (0.9), λ is the wavelength of X-ray (nm), FWHM is the full width at maxima of the peak, and θ is the Bragg angle (°).19 The average calculated particle size is 22.07 ± 3.08 nm. This value is bigger than particle size which obtained from TEM data but smaller than particle size which obtained from PSA data. However, the particle size comparison with AgNPs which synthesized by hydroxybenzoic acid or dihydroxibenzoic acid only can be performed from TEM data, because the authors did not provide the PSA and XRD data.
FTIR
The reduction of Ag+ ion to become AgNPs involved the amino and hydroxyl moieties of p-aminosalicylic acid. It can be seen from the spectra of p-aminosalicylic acid (Figure 7), the amino moiety gave strong absorption in wavenumber of 3495 and 3387 cm-1 which are twin peaks that indicate the primary amino moiety. This absorption is disappeared in the AgNPs capped by p-aminosalicylic acid spectra and covered by a wide peak of carboxylic acid. The alkene absorption around 3000 cm-1 was decreased which means that the number of double bond between carbon becomes smaller.
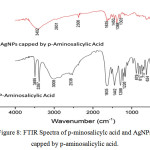 |
Figure 8: FTIR Spectra of p-minosalicylc acid and AgNPs capped by p-aminosalicylic acid. |
The Stability Evaluation of AgNPs
In order to evaluate the p-aminosalicylic acid performance as stabilizing agent, the stability of synthesized AgNPs was evaluated. The AgNPs colloid is stored in a closed bottle at normal laboratory condition. The SPR absorbance of AgNPs was observed in some time interval until 10 weeks. The colloid visually looked similar after 10 weeks of storage and the SPR spectra change can be seen in Figure 9. After 10 weeks, the wavelength number of SPR was not changed, but the absorbance changes as much as 0.137 or 7% from the initial absorbance.
Compared to the previous research, AgNPs with p-aminosalicylic acid as stabilizing agent has lower stability than AgNPs with p-hydroxybenzoic acid as stabilizing agent whose SPR absorbance change only as much as 1% from initial absorbance after 16 weeks.16 It can happen because p-aminosalicylic acid has 3 functional groups, carboxylic, amino, and hydroxyphenolic groups, whereas p-hydroxybenzoic acid only has 2 functional groups, carboxylic and hydroxyphenolic groups. The addition of amino group makes the reduction process kept going and the AgNPs become smaller but less stable.
Furthermore, the comparison also made for o,p-dihydroxibenzoic acid which also has 3 functional groups, one carboxylic and two hydroxyphenolic groups. AgNPs synthesized by p-aminosalicylic has better stabilizing performance shown by the smaller SPR absorbance change than AgNPs synthesized by o,p-dihydroxibenzoic acid whose SPR absorbance change as much as 8% after 6 weeks of storage. It was predicted that amino group has better reducing and stabilizing performance than hydroxyl group because the resulted AgNPs are smaller and more stable.
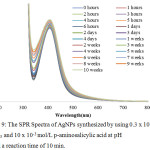 |
Figure 9: The SPR Spectra of AgNPs synthesized by using 0.3 x 10-3 mol/L AgNO3 and 10 x 10-3 mol/L p-aminosalicylic acid at pH 11 and a reaction time of 10 min. |
The Mechanism of Silver Ion Reduction and AgNPs stabilization by p-Aminosalicylic Acid
The proposed mechanism of silver ion reduction by p-aminosalicylic acid in AgNPs synthesis can be seen in Figure 10. The reduction of silver ion by p-aminosalicylic acid happened in basic condition. Beside influences p-aminosalicylic acid, basic condition also influence the silver ion. In basic condition, the H atom from hydroxyl and amino moiety of p-aminosalicylic acid was deprotonated (reaction i) and the moiety becomes more negative which enable it to react with the positive silver ion, while silver ion will aggregate to become Ag2O (reaction ii). In the water, the surface of Ag2O aggregates interacts with water and be hydrated to become AgOH which can absorb Ag+ ion (reaction iii).23 The positive silver ion which surrounds the Ag2O aggregates then interacts with the deprotonated hydroxyl and amino moiety of p-aminosalicylic acid (reaction iv). Thus, the oxidation-reduction take place with the Ag+ ion as the center of the reaction. The Ag+ ion was then reduced to become Ag0 particle which further becomes silver nanoparticles, and the deprotonated hydroxyl and amino moiety of p-aminosalicylic acid was oxidized to become keton and imine (reaction v). However, even if in the FTIR spectra of AgNPs there is absorption peak in the wavelength of 1635 cm-1, it can not verify the keton and imine formation because this absorption can also come from carboxylic moiety.
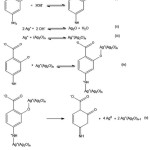 |
Figure 10: The Reduction Mechanism of Silver Ion by p-Aminosalicylic Acid. |
Beside has function to reduce the silver ion become AgNPs, p-aminosalicylic acid also stabilizes the formed AgNPs. The mechanism of AgNPs stabilization by p-aminosalicylic acid went through the repulsion force between adjacent deprotonated carboxylic acid moiety (Figure 11). This mechanism was agreed by the negative zeta potential value of the synthesized AgNPs. However, further analysis needed to confirm these proposed mechanisms. The same mechanism was also proposed by previous studies.16,17,18
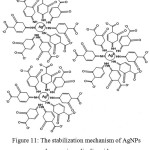 |
Figure 11: The stabilization mechanism of AgNPs by p-aminosalicylic acid. |
Conclusion
This present study reports the rapid synthesis of AgNPs from silver nitrate precursor solution with p-aminosalicylic acid as reducing and stabilizing agents. The synthesis process highly depend on the pH, reaction time, and mole ratios of the reactants. The optimum synthesis condition was pH 11, reaction time of 10 min, and a reactant mole ratio of 1:27 (AgNO3 to p-aminosalicylic acid). The formed AgNPs had been characterized by Spectrophotometry UV-Vis, TEM, PSA, XRD, and FTIR. The particle shapes of the resulting nanoparticles are spherical with a size of 32 nm. The mechanism of silver ion reduction and formed AgNPs stabilization by p-aminosalicylic acid were proposed.
Acknowledgments
The authors expressed special thanks to Ministry of Research and Higher Education of Indonesian Republic which has partly supported this research activities through Universitas Gadjah Mada, Yogyakarta, in the form of Penelitian Unggulan Perguruan Tinggi Grant and by providing scholarship of Master Education Program Leading to Doctoral Degree for Excellent Graduates (PMDSU) to the first author with contract number of 1984/UN1.P.III/DIT-LIT/LT/2017.
Conflict of Interest
All of the authors have read and agreed to this manuscript. The authors declare that there is no potential conflict of interest in this research.
References
- Fabrega, J.; Luoma, S. N.; Tyler, C. R.; Galloway, T. S.; Lead, J. R. Environ. Int. 2011, 37, 517–531.
- Rodríguez-Argüelles, M. C.; Sieiro, C.; Cao, R.; Nasi, L. J. Colloid Interface Sci. 2011, 364, 80-84.
- Prabhu, S.; Poulose, E. K. Int. Nano Lett. 2012, 32, 1-10.
- Fouda, M. M.; El-Aassar, M.; Al-Deyab, S. S. Carbohydr. Polym. 2013, 92, 1012-1017.
- Tran, Q. H.; Nguyen, V. Q.; Le, A. T. Adv. Nat. Sci. Nanosci. Nanotechnol. 2013, 4, 1-20.
- Pandey, S.; Goswami, G. K.; Nanda, K. K. Int. J. Biol. Macromol. 2012, 51, 583-589.
- Kang, C. Y.; Xi, D. L.; Chen, Y. Y.; Jiang, Z. L. Talanta. 2008, 74, 867-870.
- Dubas, S.; T Pimpan, V. Mater. Lett. 2008, 62, 2661-2663.
- Ratnarathorn, N.; Chailapakul, O.; Henry, C. S.; Dungchai, W. Talanta. 2012, 99, 552-557.
- Balakumar, V.; Prakash, P.; Muthupandi, K.; Rajan, A. Sensors Actuators, B Chem. 2017, 241, 814-820.
- Lu, Y.; Chou, K. J. Chinese Inst. Chem. Eng. 2008, 39, 673–678.
- Bin Ahmad, M.; Lim, J. J.; Shameli, K.; Ibrahim, N. A.; Tay, M. Y. Molecules. 2011, 16, 7237–7248.
- Huang, L.; Zhai, M. L.; Long, D. W.; Peng, J.; Xu, L.; Wu, G. Z.; Li, J. Q.; Wei, G. S. J. Nanoparticle Res.2008, 10, 1193–1202.
- Laudenslager, M. J.; Schiffman, J. D.; Schauer, C. L. Biomacromolecules. 2008, 9, 2682–2685.
- Hassabo, A. G.; Nada, A. A.; Ibrahim, H. M.; Abou-Zeid, N. Y. Carbohydr. Polym. 2015, 122, 343–350.
- Gusrizal, G.; Santosa, S. J.; Kunarti, E. S.; Rusdiarso, B. Int. J. ChemTech Res. 2016, 9, 472–482.
- Gusrizal, G.; Santosa, S. J.; Kunarti, E. S.; Rusdiarso, B. Asian J. Chem. 2017, 29, 1417–1422.
- Susanthy, D.; Fadliah; Wahyuni, E. T.; Santosa, S. J. Mater. Sci. Forum. 2017, 901, 26–31.
- Roto, R.; Marcelina, M.; Aprilita, N. H.; Mudasir, M.; Natsir, T. A.; Mellisani, B. Indones. J. Chem. 2017, 17, 439-445.
- Liang, A.; Liu, Q.; Wen, G.; Jiang, Z. Trac-Trend Anal. Chem. 2012, 37, 32–47.
- Susanthy, D.; Santosa, S. J.; Kunarti, E. S. Indones. J. Chem. 2018, 18, 421–427.
- Patil, R. S.; Kokate, M. R.; Jambhale, C. L.; Pawar, S. M.; Han, S. H.; Kolekar, S. S. Adv. Nat. Sci. Nanosci. Nanotechnol. 2012, 3, 1-7.
- Litvin, V. A.; Galagan, R. L.; Minaev, B. F. Colloids Surfaces A. 2012, 414, 234–243.
- Gibson, N.; Shenderova, O.; Luo, T. J. M.; Moseenkov, S.; Bondar, V.; Puzyr, A.; Purtov, K.; Fitzgerald, Z.; Brenner, D. W. Diam. Relat. Mater. 2009, 18, 620–626.
- Bin Ahmad, M.; Tay, M. Y.; Shameli, K.; Hussein, M. Z.; Lim, J. J. Int. J. Mol. Sci. 2011, 12, 4872–4884.

This work is licensed under a Creative Commons Attribution 4.0 International License.









