In Silico Docking of Phaleria Macrocarpa Bark Compounds Through Inflammation Pathway and its Cytotoxic Activities Against HCT116 Cell Line
Aryo Tedjo*1,2,3 , Fadilah Fadilah1,2,3, Kusmardi Kusmardi2,4, Rafika Indah Paramita1,2,3, Fadhilah Harmen5, Ratu Shafira Hanifah5 and Nadia Aida Novitarani5
, Fadilah Fadilah1,2,3, Kusmardi Kusmardi2,4, Rafika Indah Paramita1,2,3, Fadhilah Harmen5, Ratu Shafira Hanifah5 and Nadia Aida Novitarani5
1Departement of Medical Chemistry, Faculty of Medicine, Universitas Indonesia.
2Drug Development Researcher Center, Indonesian Medical Education and Research Institute, Faculty of Medicine, Universitas Indonesia.
3Bioinformatics Cluster, Indonesian Medical Education and Research Institute, Faculty of Medicine, Universitas Indonesia.
4Department of Anatomical Pathology, Faculty of Medicine, Universitas Indonesia, Jakarta, Indonesia.
5Medical Student of Faculty of Medicine, Universitas Indonesia.
Corresponding Author E-mail: aryo.tedjo@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/350162
Article Received on : 31-08-2018
Article Accepted on : 18-01-2018
Article Published : 19 Feb 2019
Phaleria macrocarpa (Scheff.) Boerl or Mahkota dewa in Indonesia language, a Thymelaeaceae, is widely found in Indonesia, that treat various diseases with satisfactory results. This work reports the results of in silico molecular docking of Phaleria macrocarpa bark compounds through inflammation pathway, because inflammation causes a signaling towards growth and proliferation colon cancer cell, and also testing the cytotoxic activity against HCT116 cell line. Five compounds of bark part of Phaleria macrocarpa were docked against eight inflammation protein. Bark powder of Phaleria macrocarpa was macerated in ethanol for three days, and were concentrated become bark extract. The bark extract then evaluated for cytotoxic activity on HCT116 cell line using MTT method. From the docking result, it showed that the compounds have low Gibbs energy (ΔG) in all inflammation protein. So, the compounds could inhibit the inflammation that causes a signaling towards growth and proliferation colon cancer cell. The bark extract had good IC50 value compared with Cisplatin as positive control. IC50 value of bark extract is 20.51 µg/ml. The bark extract of Phaleria macrocarpa is a potential compound to inhibit growth of HCT116 cell line.
KEYWORDS:Cytotoxicity Assay; HCT116; In Silico; Molecular Docking; Phaleria Macrocarpa
Download this article as:| Copy the following to cite this article: Tedjo A, Fadilah F, Kusmardi K, Paramita R. I, Harmen F, Hanifah R. S, Novitarani N. A. In Silico Docking of Phaleria Macrocarpa Bark Compounds Through Inflammation Pathway and its Cytotoxic Activities Against HCT116 Cell Line. Orient J Chem 2019;35(1). |
| Copy the following to cite this URL: Tedjo A, Fadilah F, Kusmardi K, Paramita R. I, Harmen F, Hanifah R. S, Novitarani N. A. In Silico Docking of Phaleria Macrocarpa Bark Compounds Through Inflammation Pathway and its Cytotoxic Activities Against HCT116 Cell Line. Orient J Chem 2019;35(1). Available from: https://bit.ly/2ttUgE0 |
Introduction
Colorectal cancer (CRC) is the one of the main reasons of most cancer-associated deaths within the international. CRC is accountable for greater than 600,000 deaths annually and occurrence charges are growing in most of the developing countries. Epidemiological and laboratory investigations advise that environmental elements consisting of western style nutritional behavior, tobacco-smoking, and absence of physical sports are taken into consideration as risks for CRC. Molecular pathobiology of CRC implicates pro-inflammatory situations to promote the tumor malignant progression, invasion, and metastasis. it’s far well known that sufferers with inflammatory bowel ailment are at higher threat of CRC. Many evidences exist reiterating the link between inflammation and CRC. infection includes interaction among numerous immune cells, inflammatory cells, chemokines, cytokines, and seasoned-inflammatory mediators, consisting of cyclooxygenase (COX) and lipoxygenase (LOX) pathways, which might also cause a signaling towards growth and proliferation cancer cell.
Phaleria macrocarpa (Scheff.) Boerl or Mahkota dewa in Indonesia language, a Thymelaeaceae, is widely found in Indonesia. Mahkota dewa has synonym of Phaleria papuana var warb wichnanmi (val) Back. In English, it is known as crown of God. Mahkota dewa fruit is frequently and empirically utilized by Indonesian to treat various diseases with satisfactory results. The bark and fruits are rich in saponins, alkaloids, polyphenolics, phenols, flavanoids, lignans and tannins. Isolated constituents of fruit include Phalerin, Icariside C3, magiferin, and gallic acid. From its seed, 29-norcurcubitacin and its derivatives, mahkoside A, mangiferin and kaempferol 3-O-β-D-glucoside had been isolated. In addition, phalerin was isolated from the methanol extract of its leave. Aglucon benzophenone and isophalerin were obtained from ethyl acetate extract of its leave. Ethyl acetate extract of its bark gave one compound of benzophenone glucoside.
The plant substances derived from the P. macrocarpa had been discovered to showcase high chemopreventive and antioxidant interest comparable to those of conventional drugs, and this activity has been attributed to the high phenolic content. P. macrocarpa is also found to have potent anti-inflammatory activity, due to its contents, including terpenoids, saponins, tannins, flavanoids and phenols such as rutin and cathecol.
This work reports the results of in silico molecular docking of Phaleria macrocarpa bark compounds (Fig.1) through inflammation pathway, because inflammation cause a signaling towards growth and proliferation cancer cell. To prove the docking result, we had extracted the bark of P. macrocarpa and further testing the cytotoxic activity against HCT116 cell line. The in vitro result then compared to in silico docking result.
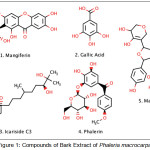 |
Figure 1: Compounds of Bark Extract of Phaleria macrocarpa. |
Experimental
Preparation of Macromolecule
3D Structure of p50 NF-κB protein (PDB id: 1SVC), Interleukin-6 protein (PDB id: 1ALU), TNF-α (PDB id: 2AZ5), Interleukin-1β (PDB id: 1ITB), PPAR-α (PDB id: 1I7G), Interleukin-2 (PDB id : 1M48), MAPK (PDB id: 3LHJ), HIF-1 (PDB id: 4H6J) were downloaded from PDB (http://www.pdb.org/). Macromolecule was prepared to calculate the binding energy using Auto Dock tools. Water and non-standard residue were removed from the protein. Hydrogen and Gasteiger partial charges were further added to the carbon that held the hydrogen. The binding pocket of the protein was determined by grid based approach using default parameters. The grid map included the binding site along with the significant portions of the surrounding surface.
Preparation of Ligand
The structure of compound was made in 2D using Marvin Sketch 15.1.19 software and saved in 3D structure in. pdb format. The ligands then being optimized using Auto Dock tools to fix the charge, added the hydrogen and minimizing energy. The 3D structure then saved in. pdbqt format.
Molecular Docking and Their Interaction Studies
Molecular Docking of designed compounds was carried out using Lamarckian genetic algorithm in Auto dock 4.2 tools with default docking parameter. We selected Auto dock4.2 tool for the purpose of molecular docking because Auto Dock has established to be powerful tool able to quick and correctly predicting certain conformations and binding energies of ligands with macro molecular targets.
The success rate in retrieving binding modes of protein–ligand complexes is an essential validation for docking programs. The measure that is commonly used to determine whether or not a binding mode prediction was successful is the RMSD (8). RMSD between the lowest energy native ligand-docked pose and native ligand-native pose was evaluated using PyMol 1.7.4.5.
Docking interactions had been clustered to decide the Gibbs energy (ΔG) and optimum docking energy conformation was considered as the fine-docked pose. The generated conformations had related value of the Gibbs energy (ΔG). An estimated inhibition concentration (Ki) was used for determination of binding energies of various docking conformations, ranking according to their binding rankings (6).
Extraction and Fractionation Samples of Seaweeds
A total of 300 g of dry powder of Phaleria macrocarpa bark are macerated (soaked) with ethanol solvent in a sealed glass vessel for 3 days, with occasional stirring. Maceration is done three times to extract as much as possible of existing substances in sample of P.macrocarpa. The mixture was then filtered; the filtrate was concentrated using rotary evaporator. Furthermore, the extract were subjected to cytotoxic activity of the extracts was evaluated as a growth inhibitor of colorectal HCT-116 cancer cells by MTT assay.
Cytotoxicity Effects
The inhibition effect of synthesized compounds against colorectal cell line, HCT116, was tested using chemical 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyl tetrazolium bromide or MTT assay. First, in 96-well plates, cells were seeded at 5000 cells/well and incubated for 24 h. Media become renewed and the cells had been added with several concentrations of the extract (6–100 µg/ml) and Cisplatin as positive control (0–16 µg/ml) incubated for a further 24 h. After 24 h, 20 µM of MTT solution (0.5 mg/ml MTT solution in media) had been added to each well and incubated for four hour at 37°C. The supernatant was aspirated and the MTT-formazan crystals formed by way of metabolically viable cells were dissolved in 100 µl of dimethyl sulphoxide (DMSO). In the end, the absorbance was monitored by a microplate reader at a wavelength of 570 nm. The percentage of viable cells was plotted versus the concentration of the test compound. The concentration by which to mediate 50% cytotoxicity (IC50) was determined by linear regression analysis.
Result and Discussion
In Silico Molecular Docking
Docking studies were performed to evaluate the effect of ligands on the various protein receptors. The result of docking simulation of compounds can be seen in table 1. Indicator from docking simulations can be seen by comparing the value of the Gibbs energy (ΔG). Gibbs energy (ΔG) showed the stability interaction between ligand and protein residues. Interaction between ligand and protein also showed in hydrogen bonds. The interaction is showed in figure 2.
Table 1: Molecular docking interaction with Inflammation Protein.
| Compound | ΔG (Kcal/Mol) | |||||||
| p50 NF-κB | IL-6 | TNF-α | IL-1β | PPAR-α | IL-2 | MAPK | HIF-1 | |
| Mangiferin | -8.6 | -8.0 | -7.4 | -10.0 | -9.5 | -7.6 | -10.7 | -7.5 |
| Gallic Acid | -5.1 | -5.2 | -4.5 | -6.2 | -5.7 | -5.0 | -6.2 | -4.3 |
| Icariside | -6.1 | -5.7 | -5.7 | -7.5 | -7.0 | -5.7 | -8.7 | -5.5 |
| Phalerin | -7.8 | -6.8 | -6.4 | -9.8 | -9.3 | -7.1 | -9.3 | -7.0 |
| Macronone | -7.9 | -7.7 | -6.9 | -8.2 | -7.3 | -7.4 | -7.7 | -7.3 |
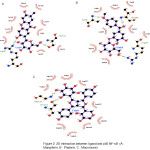 |
Figure 2: 2D interaction between ligand and p50 NF-κB (A : Mangiferin, B : Phalerin, C : Macronone). |
Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-(κ)B), is a transcription factor that regulates genes that involved in the initiation of an inflammatory response. NF-κB could be activated by any stimuli such as microbial and viral products, proinflammatory cytokines, mitokines T and B cells, also physical and chemical stresses. Expression regulation of NF-κB could be induced from any chemokines, cytokines, acute phase proteins, adhesion molecules, and antimicrobial peptides.
Briefly, most NF-κB inhibitors inhibit the activation or phosphorylation of I κB and degradation, thus preventing free NF-κB release. But approaching the direct inhibition of DNA NF-κB bonds, by interfering with the DNA binding region in NF-κB, will be more effective and can be used to design specific inhibitors. The responsible amino acids residues for DNA interaction are residue number 59-71 of the p50 subunits (Arg, Tyr, Val, Cys, Glu, Gly, Pro, Ser, His, Gly, Gly, Leu, Pro), also at residue 143, 145, 146, 243. This special p50 sequence is DBR (DNA Binding Region). From the results of the docking, we can see that the three compounds of Mahkota Dewa have an interaction on the binding site of NF-κB. So that it can inhibit the NF-κB bond with DNA, which is expected to inhibit the inflammatory response.
From the result above, it showed that the compounds have low Gibbs energy (ΔG) in all inflammation protein. So, the compounds could inhibit the inflammation that causes signaling towards growth and proliferation colon cancer cell.
Cytotoxic Assay
After completion of extraction, cytotoxic activity of bark extract was determined by MTT cell proliferation assay against HCT-116 cell line, cytotoxic activity represented by IC50. The smaller IC50 value, the higher cytotoxic activity. Anticancer activity is represented by an IC50 value (μg/mL). IC50 value <100 is considered as an active compound with anticancer activity. IC50 value in ranging from 100 to300 is considered as weak anticancer activity, whereas the IC50 value over than 300 is considered as inactive compounds. Cytotoxicity assay of bark extract and are summarized in table 2 as follow.
Table 2: Cytotoxic activity of bark extract.
| Compound | IC50 (µg/ml) |
| Extract | 20.51 ± 0.014 |
| Cisplatin |
1.35 ± 0.000 |
As shown in table 2, The IC50 value of bark extract was 20.51 μg/ml. IC50 value of extract was 20 time-fold than IC50 value of Cisplatin as positive control. From the MTT test results, it is known that the bark extract also had good cytotoxicity activity, although the IC50 was still lower than Cisplatin. But we could say that bark extract has potential activity as natural anti-colon cancer. With this research, we can look further for in vivo testing to prove the anticancer activity of Phaleria macrocarpa bark extract.
Conclusion
We described Phaleria macrocarpa for in silico and in vitro cytotoxic activities against HCT116 cell line. Compounds that contained in bark extract, showed good activity as inhibitor for inflammation protein. From cytotoxicity assay, bark extract showed good cytotoxic activity. Based on the findings of these in vitro results, further studies need to be carried out to investigate in vivo assays and toxicological studies.
Acknowledgements
We would like to thank to Universitas Indonesia for PITTA Grant 2018 that funded this research and publication.
References
- Altaf, R., Zaini, M., Dewa, A., Sadikun, A., Umar, M.I; Pharmacogn. Rev. 2013, 7(13), 73 – 80.
CrossRef - Ramdani, E.D., Marlupi, U.D., Sinambela, J., Tjandrawinata, R.R. Asian Pac. J. Trop. Biomed. 2017, 7(4), 300 – 305.
CrossRef - Shwter, A.N., et al. J. Ethnopharm. 2016, 193, 195 – 206.
CrossRef - Janakiram, N.B., Rao, C.V. Inflammation and Cancer. 2014, 25-52.
- Paramita, R.I.; Arsianti, A.; Radji, M. Intl. J. ChemTech Res. 2017, 1, 385
- Susilawati, Matsjech, S., Pranowo, H.D., Anwar, C. Indo. J. Chem. 2012, 12(1), 62 – 69.
CrossRef - Mukherjee, P.; Desai, P.; Zhou, Y. D.; Avery, M. J. Chem. Inform. Modeling. 2010, 50(5), 906–923.
CrossRef - Tor, Y. S., et al. PLoS ONE. 2015, 10(6), 1–25.
- Locatelli, C.; Filippin-Monteiro, F. B.; Creczynski-Pasa, T. B. Euro. J. Med. Chem. 2013, 60, 233–239.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









