Newly Synthesized Oxadiazole Based Mannich Base Derivatives of Fatty Acid: in Silico Study and in Vivo Anti-Hyperglycaemic Estimation
Garima Kapoor1 , Dharam Pal Pathak1, Rubina Bhutani1, Asif Husain2, Sandeep Jain3, Ravi Kant1 and Md. Azhar Iqbal2
, Dharam Pal Pathak1, Rubina Bhutani1, Asif Husain2, Sandeep Jain3, Ravi Kant1 and Md. Azhar Iqbal2
1Department of Pharmaceutical Chemistry, Delhi Institute of Pharmaceutical Sciences and Research, New Delhi, India.
2Department of Pharmaceutical Chemistry, Faculty of Pharmacy, Jamia Hamdard (Hamdard University), New Delhi, India.
3Department of Pharmaceutical Chemistry, Guru Jambeshwar University of Science and Technology, Hisar, Haryana, India.
Corresponding Author E-mail: kapoor27garima@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/340505
Article Received on : 07-06-2018
Article Accepted on : 11-08-2018
Article Published : 27 Sep 2018
A receptor peroxisome proliferator activated receptor-gamma was targeted by series of new fatty acid chemical entities (M1- M22) which was designed, synthesized and characterized by spectral analysis. Metabolites molecular properties were calculated using Lipinski’s rule of five using molinspiration online software. Docking studies were done on co-crystallized protein structure of PPAR γ, PDB-1FM9 showing M15, M17 and M8 to be best located in the active sites with scores -10.43, -10.21 and -10.00 respectively. The free binding energy estimation was done using model of Maestro 9.0 (Schrodinger) and lies between -80.15 to -61.26 kcal/mol which is significant as compared to that of standard (-48.58 Kcal/mol). Nine best docked derivatives were evaluated in-vivo for oral glucose tolerance and antihyperglycemic activity by streptozotocin induced diabetes model and M15 exhibited most promising antidiabetic activity more than the standard glibenclamide. The promising results encourage future investigation on fatty acids for development of active compounds.
KEYWORDS:Fatty acids; Hyperglycaemic; OGTT; Oleic Acid; PPAR; STZ
Download this article as:| Copy the following to cite this article: Kapoor G, Pathak D.P, Bhutani R, Husain A, Jain S, Kant R, Iqbal M.A. Newly Synthesized Oxadiazole Based Mannich Base Derivatives of Fatty Acid: in Silico Study and in Vivo Anti-Hyperglycaemic Estimation. Orient J Chem 2018;34(5) |
| Copy the following to cite this URL: Kapoor G, Pathak D.P, Bhutani R, Husain A, Jain S, Kant R, Iqbal M.A. Newly Synthesized Oxadiazole Based Mannich Base Derivatives of Fatty Acid: in Silico Study and in Vivo Anti-Hyperglycaemic Estimation. Orient J Chem 2018;34(5). Available from: http://www.orientjchem.org/?p=49984 |
Introduction
Diabetes mellitus type 2, a chronic metabolic dysfunction describes a distinctive nature of fasting hyperglycaemia. The hyperglycaemia that leads to blooming complications is an outcome of deformation of various metabolic reaction of body. Diabetes type 2 also results in disorders related to coronary, urinary tract and olfactory and eye infections. PPARs portrayed adipose tissue regulation. PPARs are target in treating diabetes mellitus, overweight also aerobic fitness.1Subtypes namely PPAR beta and gamma governs fatty acid metabolism, variety of compounds trigger them including polyunsaturated fatty acids, arachidonic acid, prostaglandins, fibrates and thiazolidinediones.2-3 Structurally different fatty acids directly act on gene expression by binding to and activating PPAR subtypes leading to decrease in blood glucose level.4
Fatty acids are very important nutritional component which are involved in cardiovascular and metabolic diseases.5 Foods containing stearic acid and other saturated fatty acids are more preferable foods for type 2 diabetic patients.6 Stimulatory action of different natural fatty acid like oleic, linoleic etc. on production of insulin in case where glucose concentration is high than in low glucose which indicates fatty acids increase glucose-stimulated insulin secretion (GSIS).7 Nitro fatty acids also have selective PPAR ligand property which is due to unique PPAR interactions of electrophilic fatty acids. These interactions encouraged us for further research due to increase in cardiovascular disease incidence in the diabetic population of recent drugs like TZD- based PPAR agonists.8 Substituted fatty acids and its derivatives mainly which have long-chain activate PPAR gamma to a certain degree and are its endogeneous ligand.9
Considering the importance and significance of fatty acid and its derivatives in treating various metabolic and other diseases in general and type II diabetes in particular, prompted us to plan the present systematic study containing oxadiazole as heterocycle hoping for the better antihyperglycemic activities than the existing ones.
Material and Methods
General
Authorized sources like Merck, Sigma-Aldrich and S.D. Fine Chemicals etc. were chosen for acquiring reagents and solvents. Thin layer chromatography on sheets of aluminium which were coated previously with silica Gel G was used to keep watch on reaction advancement at 254 nm using UV light and iodine chamber for visualization in different solvents. Open capillary method was used for resolution of melting points using Icon- Instruments which were not corrected. ATR spectrophotometer of Bruker used to indicate IR spectras. All the prepared compounds displayed their proton NMR at 300 MHz and carbon 13 NMR at 75MHz in CDCl3/DMSO-d6 as solvent on a Bruker Advance II NMR spectrophotometer. Delta values (δ) in parts per million (ppm) were used to demonstrate chemical shifts downfield in which internal standard was tetramethyl silane. The FAB mass spectrometric data (at room temperature) were measured on spectrophotometer TOF MS ES+. Perkin-Elmer 240 analyzer was taken for elemental analysis for elements Carbon, Hydrogen and Nitrogen in each derivative (range of ± 0.4%) comparing to theoretical values. The protocol for synthetic procedure and different substitutions are summarized in Scheme 1.
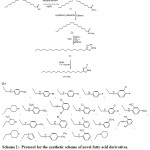 |
Scheme 1: Protocol for the synthetic scheme of novel fatty acid derivatives. |
Synthesis of methyl oleate (1)
0.25 mol of oleic acid and 1.24 mol of methanol in 100 ml of benzene were taken in a three neck 250 ml RBF on a magnetic stirrer. The ends of the condenser were added with glass tube containing Na2SO4 anhydrate and the cotton was added. To this mixture 2 ml of H2SO4 was added drop by drop through separating funnel while keeping ice around the flask with continuous shaking. When all of H2SO4 was mixed, the solution was refluxed for 5 hrs at 80°C on water bath. The product was vaporized using rotary evaporator to eliminate solvent and methanol. The residue thus obtained was mixed with 100 ml of n-hexane. This solution was washed 3 times in separating funnel with water. Water left in washing was dried by Na2SO4 anhydrous by keeping for 24hrs prior to filtering. The product was filtered to remove Na2SO4 anhydrous. Then again the residue was purified using rotary evaporator to evaporate n-hexane and gave methyl oleate as a product in liquid form. The yield of methyloleate was 91 %, m.p. 15°C and b.p. 350°C, IR: (KBr, cm-1) 2925, 2854 (C-H), 1742 (C=O), 1658 (C=C), 1169 (C-OC), 722 [(CH2)n]. 1H NMR: (CDCl3, δ, ppm): 5.42 (2H, m, CH=CH), 3.5 (3H, s, OCH3), 2.2 (2H, t, CH2), 1.96 (4H, m, 2×CH2), 1.59 (2H, m, CH2), 1.31 (20H, m, 10×CH2), 0.96 (3H, s, CH3). ESI-MS: m/z 297 (M+H). Anal. calcd. for C19H36O2: C, 76.97; H, 12.24%. Found: C, 76.84; H, 12.07%.
Synthesis of stearohydrazide (2)
RBF having 0.1mol 80% hydrazine hydrate was added with Methyl oleate (1) (0.1M) which was continuously stirred and refluxed in ethanol for 8h. The solution which was formed by the reaction was taken in a beaker and was kept on ice box. The resulting precipitate was filtered and recrystallization was done from ethanol to get stearohydrazide10. The yield was 82%, m.p. 105°C, IR: (KBr, cm-1) 3316-3100 (NH-NH2), 2921, 2853 (C-H), 1629 (O=C-NH), 1160 (C-O-C), 720 [(CH2)n]. 1H NMR: (CDCl3, δ, ppm): 8.41 (1H, s, NH), 3.66 (2H, s, NH2), 2.27 (2H, t, CH2–CONH), 1.63 (2H, m, CH2CH2–CO), 1.25 (28H, s, 14×CH2), 0.88 (3H, t, CH3). ESI-MS: m/z 299 (M+H). Anal. calcd. for C18H38N2O: C, 72.42; H, 12.83; N, 9.38%. Found: C, 72.80; H, 12.65; N, 9.14%.
Synthesis of 5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (3)
RBF filled with stearohydrazide (0.038 mol) was added up with absolute ethanol (40 mL) to dissolve it. 0.034 mol of carbon disulfide (2 mL) was then mixed with solution following with the addition of a solution of 0.019 mol of potassium hydroxide in water (20 mL). Mixture obtained was refluxed for 11hrs with continuous stirring. Initial yellow solution changed to green and after that it got converted to green and then at last into light yellow when the reaction proceeds. Hydrogen sulphide gas was evolved during the progress of reaction. When the reaction is being executed it was taken to room temperature and allowed to cool and ethanol was vaporized. Mixture was diluted with distilled water (200 ml) and clabber to pH 2-3 by 4N HCl. It was then strained, flushed with ether and recrystallized with ethanol.11 The yield was 64 %, m.p. 150oC, IR: (KBr, cm-1) 2920, 2851 (C-H), 3314 (N-H), 1602 (C=N), 1306 (N-N=C), 1163 (C=S), 1067 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 7.2 (s, NH), 1.32 (4H, m, 2×CH2), 1.27 (26H, m, 13×CH2), 0.94 (3H, t, CH3). ESI-MS: m/z 341 (M+H). Anal. calcd. for C19H36N2OS: C, 67.01; H, 10.65; N, 8.23%. Found: C, 67.33; H, 10.93; N, 8.11%.
Typical Synthetic procedure of 3-Substituted aminomethyl-5-heptadecyl-1,3,4-oxadiazole-2-thiones (M1-M22)
In a beaker having formaldehyde (37%, 0.05 mol), solution of 3 (0.01 mol) in 1:1 ratio of dioxane and absolute ethanol (20ml) was added. To this mixture, an appropriate amine (0.01M) dissolved in 10ml of alcohol (10 mL) was mixed with continuous overnight stirring on Parallel synthesizer. The mannich bases were precipitated when the resulting solution was added to crush ice. The precipitates were filtered, dried and crystallization was done by ethanol to get synthetic derivatives M1-M22.12-13
5-heptadecyl-3-((4-nitrophenylamino)methyl)-1,3,4-oxadiazole-2(3H)-thione (M1)
Yield: 49%, m.p.: 285–287°C, IR: (KBr, cm-1) 2930 (N-H), 1618 (C=N), 1306 (N-N=C), 1237 (C=S), 1067 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.86 (3H, t, CH3), 1.25-1.33 (28H, m, 14xCH2), 1.54 (2H, m, CH2), 2.14 (2H, t, CH2), 4.91 (2H, s-CH2-NH), 5.94(1H, s, -CH2-NH), 6.89 [2H, m, 2xCH-(Ar)], 8.12 [2H, m, 2xCH-(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.04, 22.26, 25.59, 26.60, 29.70, 29.46, 30.20, 32.49, 56.90, 117.36, 118.16, 140.30, 140.55, 155.81, 175.19. ESI-MS: m/z 492 (M+H); Anal. calcd. for C26H42N4O3S: C, 63.64; H, 8.63; N,11.42%. Found: C, 63.45; H, 8.82; N, 11.55%.
3-((4-bromophenylamino)methyl)-5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (M2)
Yield: 53%, m.p.: 280–282°C, IR: (KBr, cm-1) 3001 (N-H), 1618 (C=N), 1306 (N-N=C), 1237 (C=S), 1067 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.88 (3H, t, CH3), 1.15-1.35 (28H, m, 14xCH2), 1.56 (2H, m, CH2), 2.21(2H, t, CH2), 4.72 (2H, s, CH2-NH), 5.81 (1H, s, -CH2-NH), 6.73 [2H, m, 2xCH-(Ar)], 7.91 [2H, m, 2xCH-(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.20, 22.16, 25.99, 26.30, 29.70, 29.24, 30.50, 32.49, 57.17, 117.29, 122.31, 132.80, 140.10, 155.18, 175.49. ESI-MS: m/z 525 (M+H). Anal. calcd. for C26H42BrN3OS: C, 59.53; H, 8.07; N, 8.01%. Found: C, 59.31; H, 8.28; N, 8.33%.
3-((4-chlorophenylamino)methyl)-5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (M3)
Yield: 61%, m.p.: 282–284°C, IR: (KBr, cm-1) 3012 (N-H), 1620 (C=N), 1299 (N-N=C), 1204 (C=S), 1100 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.86 (3H, t, CH3), 1.11-1.37 (28H, m, 14xCH2), 1.53 (2H, m, CH2), 2.61 (2H, t, CH2), 4.69 (2H, s, -CH2-NH), 5.93 (1H, s, -CH2-NH), 6.61 [2H, m, 2xCH-(Ar)], 7.42 [2H, m, 2xCH-(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 13.10, 22.16, 25.91, 26.70, 28.40, 29.49, 30.50, 32.24, 57.77, 119.18, 129.42, 129.19, 140.80, 155.48, 175.79. ESI-MS: m/z 480 (M+H). Anal. calcd. for C26H42ClN3OS: C, 65.04; H, 8.82; N, 8.75%. Found: C, 65.24; H, 8.94; N, 8.62%.
3-((4-fluorophenylamino) methyl)-5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (M4)
Yield: 48%, m.p.: 275–277°C, IR: (KBr, cm-1) 3009 (N-H), 1640 (C=N), 1260 (N-N=C), 1234 (C=S), 1021 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.89 (3H, t, CH3), 1.14-1.37 (28H, m, 14xCH2), 1.59 (2H, m, CH2), 2.51 (2H, t, CH2), 4.75 (2H, s, -CH2-NH), 5.93 (1H, s, -CH2-NH), 6.87 [2H, m, 2xCH-(Ar)], 6.99 [2H, m, 2xCH-(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.06, 22.66, 25.89, 26.40, 29.99, 30.50, 32.48, 57.87, 117.10, 118.87, 140.90, 155.08, 160.51, 175.89.ESI-MS: m/z 464 (M+H). Anal. calcd. for C26H42FN3OS: C, 67.35; H, 9.13; N, 9.06%. Found: C, 67.29; H, 9.05; N, 9.34%.
3-((3-chloro-4-fluorophenylamino)methyl)-5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (M5)
Yield: 58%, m.p.: 288–290°C, IR: (KBr, cm-1) 2950 (N-H), 1623 (C=N), 1306 (N-N=C), 1163 (C=S), 1067 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.93 (3H, t, CH3), 1.15-1.37 (28H, m, 14xCH2), 1.60 (2H, m, CH2), 2.61 (2H, t, CH2), 4.69 (2H, s, -CH2-NH), 5.93 (1H, s, -CH2-NH), 6.31 [1H, m, 2xCH-(Ar)], 7.21 [1H, m, CH-(Ar)], 7.42 [1H, m, CH-(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.90, 22.16, 25.29, 26.06, 29.90, 30.90, 32.14, 57.47, 116.57, 118.77, 119.91, 140.80, 152.21, 155.48, 175.89. ESI-MS: m/z 498 (M+H). Anal. calcd. for C26H42ClFN3OS: C, 62.69; H, 8.30; N, 8.44%. Found: C, 62.36; H, 8.53; N, 8.22%.
5-heptadecyl-3-(piperazin-1-ylmethyl)-1,3,4-oxadiazole-2(3H)-thione (M6)
Yield: 61%, m.p.: 281–283°C, IR: (KBr, cm-1) 3052 (N-H), 1613 (C=N), 1302 (N-N=C), 1120 (C=S), 1084 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.86 (3H, t, CH3), 1.14-1.47 (32H, m, 16xCH2), 2.34 (1H, s, NH of piperazine), 2.53-2.73 [8H, m, 4xCH2of piperazine], 3.66 (2H, s, N-CH2-N). 13C NMR: (DMSO-d6, δ, ppm): 14.09, 22.66, 25.29, 26.50, 29.66, 29.49, 31.80, 32.4, 45.86, 50.79, 64.30, 155.48, 175.29. ESI-MS: m/z 439 (M+H). Anal. calcd. for C24H46N4OS: C, 65.71; H, 10.57; N, 12.77%. Found: C, 65.63; H, 10.85; N, 12.99%.
3-((2,4-dinitrophenylamino)methyl)-5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (M7)
Yield: 38%, m.p.: 290–292°C, IR: (KBr, cm-1) 3061 (N-H), 1604 (C=N), 1309 (N-N=C), 1161 (C=S), 1108 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.87 (3H, t, CH3), 1.05-1.25 (28H, m, 14xCH2), 1.58 (2H, t, CH2), 2.26 (2H, t, CH2), 5.33 (2H, s, -CH2-NH), 6.63 (1H, s, -CH2-NH), 8.2 [1H, m, CH-(Ar)], 9.10-9.12 [2H, m, 2xCH-(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.20, 22.36, 25.49, 26.70, 29.10, 29.46, 30.50, 32.94, 57.67, 103.84, 117.83, 132.59, 140.05, 155.18, 175.93. ESI-MS: m/z 341 (M+H). Anal. calcd. for C26H41N5O5S: C, 58.29; H, 7.71; N, 13.07%. Found: C, 58.48; H, 7.52; N, 13.13%.
3-((2,6-dichlorophenylamino)methyl)-5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (M8)
Yield: 48%, m.p.: 296–298°C, IR: (KBr, cm-1) 2920, 2851 (N-H), 1622 (C=N), 1300 (N-N=C), 1163 (C=S), 1065 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.87 (3H,t,CH3), 1.13-1.39 (28H, m, 14xCH2), 1.57 (2H, m, CH2), 2.51 (2H, t, CH2), 4.73 (2H, s, -CH2-NH), 5.6 (1H, s, -CH2-NH), 7.26 [1H, m, CH-(Ar)], 7.47 [2H, m, 2xCH-(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.10, 22.67, 25.69, 27.20, 29.30, 29.64, 31.91, 32.4, 56.95, 77.43, 113.09, 126.09, 140.43, 150.01, 163.03, 177.14.ESI-MS: m/z 515 (M+H). Anal. calcd. for C26H41Cl2N3OS: C, 60.68; H, 8.03; N, 8.17%. Found: C, 60.44; H, 8.35; N, 8.48%.
3-((2,4-dichlorophenylamino)methyl)-5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (M9)
Yield: 67%, m.p.: 285–287°C, IR: (KBr, cm-1) 2928 (C-H), 1617 (C=N), 1306 (N-N=C), 1237 (C=S), 1102 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.86 (3H, t, CH3), 1.10-1.37 (28H, m, 14xCH2), 1.52 (2H, m, CH2), 2.58 (2H, t, CH2), 4.51 (2H, s, -CH2-NH), 6.09 (1H, s, -CH2-NH), 6.88 [1H, m, 2xCH-(Ar)], 7.35 [1H, m, CH-(Ar)], 7.76 [1H, m, CH-(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.66, 22.65, 25.91, 26.30, 29.80, 30.77, 32.14, 56.97, 117.13, 122.27, 128.68, 129.22, 130.42, 135.90, 155.18, 175.79. ESI-MS: m/z 515 (M+H). Anal. calcd. for C26H41Cl2N3OS: C, 60.68; H, 8.03; N, 8.17%. Found: C, 60.49; H, 8.24; N, 8.05%.
3-((3,4-dichlorophenylamino)methyl)-5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (M10)
Yield: 63%, m.p.: 280–282°C, IR: (KBr, cm-1) 2932 (N-H), 1668 (C=N), 1311 (N-N=C), 1152 (C=S), 1054 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.87 (3H, t, CH3), 1.18-1.37 (28H, m, 14xCH2), 1.53 (2H, m, CH2), 2.66 (2H, t, CH2), 4.89 (2H, s, -CH2-NH), 5.80 (1H, s, -CH2-NH), 6.67 [1H, m, 2xCH-(Ar)], 7.36 [1H, m, CH-(Ar)], 7.88 [1H, m, CH-(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.51, 22.61, 25.19, 26.77, 29.10, 29.49, 30.90, 32.44, 57.97, 119.58, 121.15, 125.58, 130.54, 131.68, 140.70, 155.18, 175.93. ESI-MS: m/z 515 (M+H). Anal. calcd. for C26H41Cl2N3OS: C, 60.68; H, 8.03; N, 8.17%. Found: C, 60.49; H, 8.35; N, 8.29%.
5-heptadecyl-3-((2-methyl-4-nitrophenylamino)methyl)-1,3,4-oxadiazole-2(3H)-thione (M11)
Yield: 49%, m.p.: 280–282°C, IR: (KBr, cm-1) 2932 (N-H), 1613 (C=N), 1315 (N-N=C), 1169 (C=S), 1020 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.83 (3H, t, CH3), 1.22-1.39 (32H, m, 16xCH2), 2.34 (3H, m, Ar-CH3), 4.51 (2H, s, -CH2-NH), 5.74 (1H, s, -CH2-NH), 6.52-8.02 [3H, m, 3xCH-(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 17.17, 22.66, 25.19, 26.80, 29.19, 29.99, 32.42, 57.79, 116.54, 120.12, 127.92, 140.59, 155.68, 175.83. ESI-MS: m/z 505 (M+H). Anal. calcd. for C27H44N4O3S: C, 64.25; H, 8.79; N, 11.10%. Found: C, 64.32; H, 8.62; N, 11.41%.
5-heptadecyl-3-((2-methylpiperazin-1-yl)methyl)-1,3,4-oxadiazole-2(3H)-thione (M12)
Yield: 71%, m.p.: 297–299°C, IR: (KBr, cm-1) 2919 (N-H), 1629 (C=N), 1225 (N-N=C), 1173 (C=S), 1040 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.86 (3H, t, CH3), 1.17 (3H, d, CH3 of piperazine), 1.29-1.47 (32H, m, 16xCH2), 2.14 (1H, s, NH of piperazine), 2.55-2.78 [6H, m, 3xCH2 of piperazine], 3.08 (1H, m, CH of piperazine), 3.76 (2H, s, N-CH2-N). 13C NMR: (DMSO-d6, δ, ppm): 14.53, 16.65, 22.63, 25.91, 26.90, 29.80, 29.49, 30.88, 32.49, 45.67, 47.08, 50.91, 58.73, 65.60, 155.81, 175.93. ESI-MS: m/z453 (M+H). Anal. calcd. for C25H48N4OS: C, 66.32; H, 10.69; N, 12.38%. Found: C, 66.15; H, 10.77; N, 12.07%.
3-((p-toluidino)methyl)-5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (M13)
Yield: 51%, m.p.: 274–276°C, IR: (KBr, cm-1) 2920 (N-H), 1660 (C=N), 1335 (N-N=C), 1192 (C=S), 1063 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.85 (3H, t, CH3), 1.12-1.26 (28H, m, 14xCH2), 1.57 (2H, t, CH2), 2.18 [3H, s, CH3(Ar)], 2.56 (2H, t, CH2), 4.78 (2H, s, CH2-NH), 5.45 (1H, s, -CH2-NH), 6.86 [2H, m, 2xCH(Ar)], 7.06 [2H, m, 2xCH(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.0, 21.3, 22.6, 25.9, 26.0, 29.0, 29.4, 32.4, 57.7, 116.9, 129.6, 132.5, 140.99, 155.78, 175.91. ESI-MS: m/z 461 (M+H). Anal. calcd. for C27H45N3OS: C, 70.54; H, 9.87; N, 9.14%. Found: C, 70.76; H, 9.68; N, 9.41%.
3-((2-bromophenylamino)methyl)-5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (M14)
Yield: 57%, m.p.: 281–283°C, IR: (KBr, cm-1) 2971 (N-H), 1618 (C=N), 1289 (N-N=C), 1201 (C=S), 1061 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.91 (3H, t, CH3), 1.13-1.45 (28H, m, 14xCH2), 1.54 (2H, t, CH2), 2.58 (2H, t, CH2), 4.71 (2H, s, -CH2-NH), 5.92 (1H, s, -CH2-NH), 6.31 [1H, m, CH-(Ar)], 6.91-7.01 [2H, m, 2xCH-(Ar)], 7.34 [1H, m, CH-(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.11, 22.66, 25.19, 26.09, 29.30, 29.47, 30.77, 32.44, 57.17, 113.33, 122.07, 126.73, 128.24, 132.43, 135.42, 155.78, 175.98. ESI-MS: m/z 525 (M+H). Anal. calcd. for C26H42BrN3OS: C, 59.53; H, 8.07; N, 8.01%. Found: C, 59.72; H, 8.18; N, 8.33%.
3-((2-fluorophenylamino)methyl)-5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (M15)
Yield: 62%, m.p.: 298–300°C, IR: (KBr, cm-1) 3009 (N-H), 1609 (C=N), 1304 (N-N=C), 1154 (C=S), 1063 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.89 (3H, t, CH3), 1.15-1.35 (28H, m, 14xCH2), 1.59 (2H, t, CH2), 2.56 (2H, t, CH2), 4.63 (2H, s, -CH2-NH), 5.91 (1H, s, -CH2-NH), 6.81 [1H, m, CH-(Ar)], 6.92-7.06 [3H, m, 3xCH-(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.55, 22.16, 25.95, 26.70, 29.03, 29.14, 30.90, 32.43, 57.37, 115.67, 121.82, 125.21, 126.83, 152.96, 155.81, 175.59. ESI-MS: m/z464 (M+H). Anal. calcd. for C26H42FN3OS: C, 67.35; H, 9.13; N, 9.06%. Found: C, 67.65; H, 9.04; N, 9.24%.
5-heptadecyl-3-((3-methoxyphenylamino)methyl)-1,3,4-oxadiazole-2(3H)-thione (M16)
Yield: 55%, m.p.: 301–303°C, IR: (KBr, cm-1) 2988 (N-H), 1607 (C=N), 1309 (N-N=C), 1165 (C=S), 1067 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.83 (3H, t, CH3), 1.25-1.39 (32H, m, 16xCH2), 3.44 (3H, m, O-CH3), 4.31 (2H, s, -CH2-NH), 4.96 (1H, s, -CH2-NH), 5.42-6.92 [4H, m, 4xCH-(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.04, 22.65, 25.91, 26.60, 29.09, 29.14, 30.40, 32.49, 57.67, 106.27, 108.13, 119.21, 130.45, 140.66, 155.18, 160.39, 175.93. ESI-MS: m/z 476 (M+H). Anal. calcd. for C27H45N3O2S: C, 68.17; H, 9.53; N, 8.83%. Found: C, 68.35; H, 9.62; N, 8.96%.
3-((2,6-dimethylphenylamino)methyl)-5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (M17)
Yield: 69%, m.p.: 288–290°C, IR: (KBr, cm-1) 2922 (N-H), 1622 (C=N), 1347 (N-N=C), 1163 (C=S), 1082 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.95 (3H, t, CH3), 1.15-1.26 (28H, m, 14xCH2), 1.54 (2H, m, CH2), 1.73 (2H, m, CH2), 2.11-2.67 [6H, s, 2xCH3(Ar)], 2.27 (2H, t, CH2), 5.34 (2H, s, CH2-NH), 5.9 (1H, s, -CH2-NH), 7.02-7.05 [1H, t, CH(Ar)], 7.26 [2H, m, 2xCH(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.10, 18.12, 22.63, 25.79, 26.99, 29.03, 29.45, 30.66, 32.74, 57.79, 126.87, 129.59, 129.99, 140.91, 155.86, 175.91. ESI-MS: m/z 475 (M+H). Anal. calcd. for C28H47N3OS: C, 70.99; H, 10.00; N, 8.87%. Found: C, 70.74; H, 10.22; N, 8.99%.
5-heptadecyl-3-(morpholinomethyl)-1,3,4-oxadiazole-2(3H)-thione (M18)
Yield: 67%, m.p.: 296–298°C, IR: (KBr, cm-1) 2960 (N-H), 1602 (C=N), 1351 (N-N=C), 1201 (C=S), 1099 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.86 (3H, t, CH3), 1.04-1.67 (32H, m, 16xCH2), 2.73-2.83 [8H, m, 4xCH2 of morpholine], 3.56 (2H, s, N-CH2-N). 13C NMR: (DMSO-d6, δ, ppm): 14.10, 22.16, 25.29, 26.90, 29.09, 29.49, 30.90, 32.46, 50.12, 65.50, 66.76, 155.88, 175.96. ESI-MS: m/z 440 (M+H). Anal. calcd. for C24H45N3O2S: C, 65.56; H, 10.32; N, 9.56%. Found: C, 65.42; H, 10.64; N, 9.38%.
3-((1H-imidazol-1-yl)methyl)-5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (M19)
Yield: 48%, m.p.: 290–292°C, IR: (KBr, cm-1) 2998 (N-H), 1605 (C=N), 1309 (N-N=C), 1155 (C=S), 1019 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.93 (3H, t, CH3), 1.14-1.33 (32H, m, 16xCH2), 4.56 (2H, s, N-CH2-N), 6.55-7.3 [(3H, m, 3xCH(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.21, 22.26, 25.92, 26.09, 29.90, 30.90, 32.74, 65.00, 118.31, 131.51, 136.66, 155.18, 175.49. ESI-MS: m/z 421 (M+H). Anal. calcd. for C23H40N4OS: C, 65.67; H, 9.58; N, 13.32%. Found: C, 65.44; H, 9.49; N, 13.63%.
3-((4-ethylphenylamino)methyl)-5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (M20)
Yield: 58%, m.p.: 282–284°C, IR: (KBr, cm-1) 2987 (N-H), 1618 (C=N), 1306 (N-N=C), 1167 (C=S), 1055 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.93 (3H, t, CH3), 1.15-1.39 (32H, m, 16xCH2), 1.44 (3H, m, Ar-CH2-CH3), 1.81 (2H, m, Ar-CH2-CH3), 4.61 (2H, s, -CH2-NH), 5.74 (1H, s, -CH2-NH), 6.42-7.12 [4H, m, 4xCH-(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.17, 22.63, 25.87, 26.80, 28.50, 29.01, 29.47, 30.80, 32.48, 40.14, 57.97, 116.19, 129.56, 140.70, 151.27, 155.38, 175.49. ESI-MS: m/z 474 (M+H). Anal. calcd. for C28H47N3OS: C, 70.99; H, 10.00; N, 8.87%. Found: C, 70.85; H, 10.22; N, 8.79%.
5-heptadecyl-3-((2-nitrophenylamino)methyl)-1,3,4-oxadiazole-2(3H)-thione (M21)
Yield: 61%, m.p.: 286–288°C, IR: (KBr, cm-1) 2990 (N-H), 1598 (C=N), 1318 (N-N=C), 1146 (C=S), 1061 (C-O-C); 1H NMR: (CDCl3, δ, ppm): 0.89 (3H, t, CH3), 1.15-1.39 (28H, m, 14xCH2), 1.51 (2H, m, CH2), 2.04 (2H, t, CH2), 4.41 (2H, s, -CH2-NH), 5.94 (1H, s, -CH2-NH), 6.69-8.12 [4H, m, 4xCH-(Ar)]. 13C NMR: (DMSO-d6, δ, ppm): 14.03, 22.63, 25.96, 26.09, 29.08, 29.49, 30.33, 32.88, 58.79, 117.37, 117.15, 122.27, 127.67, 132.89, 140.55, 155.98, 175.91. ESI-MS: m/z 491 (M+H). Anal. calcd. for C26H42N4O3S: C, 63.64; H, 8.63; N, 11.42%. Found: C, 63.52; H, 8.45; N, 11.23%.
5-heptadecyl-3-(piperidin-1-ylmethyl)-1,3,4-oxadiazole-2(3H)-thione (M22)
Yield: 64%, m.p.: 280–282°C, IR: (KBr, cm-1) 2914 (N-H), 1630 (C=N), 1298 (N-N=C), 1199 (C=S), 1020 (C-O-C). 1H NMR: (CDCl3, δ, ppm): 0.82 (3H,t,CH3), 1.45 (2H, m, CH2 of piperidine), 1.14-1.71 (30H, m, 16xCH2), 2.52 (2H, t, CH2), 2.43-2.83 [8H, m, 4xCH2(Ar)], 4.2 (2H, s, N-CH2-N). 13C NMR: (DMSO-d6, δ, ppm): 14.12, 22.46, 24.61, 25.89, 26.01, 29.09, 29.99, 32.49, 52.98, 64.90, 155.68, 175.19.ESI-MS: m/z 438 (M+H). Anal. calcd. for C25H47N3OS: C, 68.60; H, 10.82; N, 9.60%. Found: C, 68.98; H, 10.91; N, 9.43%.
In Silico Computational Studies
Adsorption, Distribution, Metabolism and Excretion Property Prediction: Lipinski screening test
Drug-likeness of the compounds was analyzed through online Molinspiration property calculation toolkit.14 Various pharmacokinetic properties of all the synthesized compounds were determined using Lipinski’s rule like TPSA, molecular weight, log P, hydrogen bond donors and acceptors; rotatable bonds and molecular volume. This rule states compound to not have greater than 5 donor and 10 acceptor of hydrogen bond, molecular weight to be less than 500 dalton and 5 or less than 5 of coefficient of partition between octanol and water. Other physicochemical parameter i.e. Topological Polar Surface Area (TPSA) of all synthesized derivatives was deliberated to notice deficiently absorbed analogs having reduced CNS bioavailability helping in selection orally active drugs. Candidate with TPSA >60Å are basically chosen.15-18 %ABS (% Ab) of all the derivatives was calculated from the value of TPSA by applying the formula:
%Ab = 109 – [0.345 ×TPSA]
Molecular docking and Prime MMGBSA19
Computations of all synthetic derivatives were carried by Schrodinger programme Maestro 9.0, New York, USA on PDB ID-1FM9 which is PPAR-γ crystal taken from RCSB. The interpretations of different means of binding of new entities with cavities of the receptor were done. The well demonstrated aim in anti-hyperglycaemic action is PPAR gamma receptor. Protein data bank (PDB 1FM9) was opened and the basic crystal structure was downloaded. Tool named Lig prep was used in preparing ligands ie. new chemical derivatives and protein preparation wizard in preparation of structure of protein. Molecules of water not desired were eliminated physically by software from the framework. The structure of protein was intensified and then its energy was minimized by root-mean-square deviation of 0.3 Å. Ligands energy was also minimized with the OPLS 2005 force field. Glidescores and free binding energy of docked compounds was used in evaluating binding affinities.
Software Maestro 9.0 is used to estimate binding energy of the ligands i.e. synthetic analogs and glibenclamide against receptor having PDB ID 1FM9 which is crystal structure of PPAR γ through method called as Prime molecular mechanics-generalized born surface area. Conclusions can be obtained by regulating MM-GBSA program instantly from the file created by docking procedure. The perfect organization and binding resemblance of the novel entities on the vital positions of the receptor with the nearby amino acids are anticipated by docking glidescore, free binding energy,H bond and pi-pi bonding. The protein-ligand energy in coulomb-vdW is elucidated by the Emodel function.
Pharmacology
Rats (Albino wistar, male/female) of weight 200-250gm were taken in our research work acquired from Animal House centre of DIPSAR, New Delhi. They were housed in polypropylene cages at normal laboratory condition (temperature 22±2oC and relative humidity 45 ± 5% with 12h day: 12h night cycle). All animals were given water ad libitum and normal pellet diet. The study was executed after taking approval of Institutional Animal Ethical Committee (IAEC) through protocol IAEC ⁄ 2016-I/ Prot. No.05 according to CPCSEA guidelines. Animals were deprived of food for at least 18 hrs but were allowed free access to drinking water.21-22 Blood glucose level was deliberated by automatic machine Accu-Chek Active Glucose by Roche Diagnostics.
Effects on Oral glucose tolerance in Normal Rats
M2, M4, M8, M9, M12, M13, M15, M17 and M20 were selected for OGTT through their docking scores in molecular modeling studies. Overnight (16hrs) fasted normal rats were used in OGTT. 11 groups were made; six rats were kept in each group. Group 1 (Normal group) was given 0.5% weight by volume aqueous CMC solution (5 mL/kg) orally. Group 2 (Standard group) was treated with glibenclamide 10 mg/kg in 0.5% weight by volume aqueous CMC. Groups 3–11 received test derivatives (100 mg/kg of body weight) by oral gavage. 30 min after giving of CMC, glibenclamide and test entities, 5g/kg of body weight of glucose was given orally. Blood glucose level was measured at 0h (just before), 30min and 90min of orally given synthetic derivatives by Roche Accu-chek Active TM Test meter by its strips through retro orbital plexus of rat eye.23-24
Effects on streptozotocin-induced diabetic rats25-28
Dose of 10 mg/kg body weight of Glibenclamide prepared 0.5% weight by volume aqueous CMC was served to rats as a reference standard. Rats were refrained for a night and diabetes was initiated by injection of one dose of a newly prepared STZ in 0.1M citrate buffer (60 mg/kg body weight) intraperitoneally. Streptozotocin induced lowering of glucose level was overcomed by giving 5% glucose solution for a night to all rats. Blood glucose level was measured by glucose meter after 48hrs. Rats with 250 mg/dL and more glucose level were separated and considered as diabetic which were used for further study.21
Group design
Compounds M2, M4, M8, M9, M12, M13, M15, M17 and M20 were selected for anti-diabetic potential in STZ diabetic rat model through glidescores. Eleven groups were made with six rats in each. Group 1 (Diabetic control group) was given 0.5% weight by volume aqueous CMC solution (5 mL/kg body weight) orally. Group 2 (Standard group) was served with 10 mg/kg glibenclamide,0.5% weight by volume aqueous CMC. Groups 3–11 (synthetic analog groups) received synthetic derivatives at a dose of 100 mg/kg body weight orally through oral gavage.
Blood Glucose Measurement
Under mild ether anaesthesia blood was taken by retro orbital plexus of eye of rat using capillary tube. The quantity of sugar level of blood in each animal was resoluted after 0, 2, 4, 6 hours by Test strips of Roche apparatus named Accu-chek Active TM Test meter.
Statistical analysis for OGTT and STZ induced diabetic rats
Results for OGTT in normal and STZ in induced diabetic rats are given as means ± SEM (n=6). Difference in statistical data of synthesized and normal was done through one-way ANOVA then dunnett’s multiple comparison tests. The data accounted as significant at p<0.05 and p<0.01.
Investigation of Results
Novel derivatives of fatty acid as PPAR agonists were designed and synthesized following Scheme 1. Initially 5-heptadecyl-1,3,4-oxadiazole-2(3H)-thione (3) is needed as starting material got by ester formation reaction of oleic acid with methanol emerged into methyl oleate (1), refluxed escorted by hydrazine hydrate in ethanol to provide stearohydrazide (2) and (2) was then reacted with CS2 and KOH in ethanol to give (3). Similar type of substituted anilines was acted on the compound 3 to yield the respective N-Mannich bases M1-M22. The structures of synthesized compounds M1-M22 were established by C, H, N estimation, and proton and carbon nuclear magnetic resonance and also by MASS spectroscopy. The IR spectra of synthesized derivatives showed absorption bands, NH bands at 2851–3432cm-1 and C= S stretching band at 1120–1237cm-1. The 1H NMR spectrum showed singlet at 3.66-5.34 parts per million merging for protons of –N–CH2–N and its δ was in range of 57.1-58.79 parts per million by 13C NMR spectra. Synthesized entities showed weak molecular ion peaks showing instability of molecular ions. Range of ±0.4% compared to theoretical values elements (C, H, N) were obtained by elemental analysis.
Computational Study
Screening of Pharmacokinetic Properties
ADME property prediction results of synthesized fatty acid derivatives (M1-M22) were deliberated online using Molinspiration software given in Table 1. The range of absorption percentage (% ABS) was from 62.54 to 97.20 for the derivatives indicating that these compounds have good permeability in the cellular membrane. The values of TPSA of novel compounds were endowed in range of 34.20–134.64Å captivating secondary constructional up gradation for the development of novel entities.
Table 1: ADME property estimation.
|
Compd.Code |
%ABS |
TPSAa |
n-rotbb |
Molecular Weight |
Molecular Volume |
Log Pc |
nOHNHd |
None |
|
M1 |
78.35 |
88.82 |
20 |
490.7 |
484.02 |
8.96 |
1 |
7 |
|
M2 |
94.17 |
|||||||
|
M3 |
94.17 |
42.99 |
19 |
524.6 |
478.57 |
9.24 |
1 |
4 |
|
M4 |
94.17 |
|||||||
|
M5 |
94.17 |
42.99 |
19 |
480.16 |
474.22 |
9.2 |
1 |
4 |
|
M6 |
93.05 |
|||||||
|
M7 |
62.54 |
42.99 |
19 |
463.71 |
465.62 |
9.03 |
1 |
4 |
|
M8 |
94.17 |
|||||||
|
M9 |
94.17 |
42.99 |
19 |
498.15 |
479.15 |
9.23 |
1 |
4 |
|
M10 |
94.17 |
|||||||
|
M11 |
78.35 |
46.23 |
18 |
438.73 |
458.43 |
7.48 |
1 |
5 |
|
M12 |
93.05 |
|||||||
|
M13 |
94.17 |
134.64 |
21 |
535.71 |
507.35 |
8.91 |
1 |
10 |
|
M14 |
94.17 |
|||||||
|
M15 |
94.17 |
42.99 |
19 |
514.59 |
487.76 |
9.36 |
1 |
4 |
aTopological polar surface area (TPSA). bNumber rotatable bonds (n-rotb). cLog of partition coefficient n-octanol/water (LogP). dHydrogen bond donors (nOHNH). eHydrogen bond acceptor (nON).
Docking Studies
Molecular Docking was done to get report of the binding motif of new fatty acid entities with the active region of PPAR-γ crystal form with PDB ID-1FM9. This study was done by Schrodinger programme Maestro 9.0. Glidescores of docking study and free binding energies of all anlogs are presented in Table 2. M15, M17 and M8 showed highest docking score (-10.43, -10.21 and -10.00) and located well in PPAR-γ vital positions when compared with standard drug (-10.41). Analysis of the compound M15 binding mode in the active site revealed that oxygen atom of oxadiazole ring showed three hydrogen bonds with three different amino acids SER 289, HIP 323 and TYR 473 respectively (Fig. 1). The significance of oxadiazole ring for reticence and obligatory effectiveness is underscored by these interactions with PPAR-γ. Figures (Fig. 2 and Fig. 3) showed that docking mode of compound M17 and M8 showed identical three hydrogen bonds interaction. Out of three, two hydrogen bondswere observed between oxygen atom of oxadiazole ring and amino acid HIP 323 and TYR 473 whereas there was one more H-bond between -NH and MET 364. Elaborated binding patterns of well docked analogs (M15, M17 and M8 within 3.5Å) are shown in figures.
 |
Figure 1: 3D interrelationship picture of M15 (PDB ID: 1FM9) presenting hydrogen bonds with yellow dot lines in relation to amino acid SER 289, HIP 323 and TYR 473 respectively. |
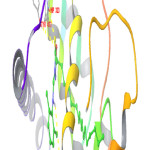 |
Figure 2: 3D interrelationship picture of M17 (PDB ID: 1FM9) presenting hydrogen bonds with yellow dot lines in relation to amino acid MET 364, HIP 323 and TYR 473 respectively. |
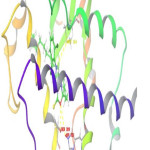 |
Figure 3: 3D interrelationship picture of M8 (PDB ID: 1FM9) presenting hydrogen bonds with yellow dot lines in relation to amino acid MET 364, HIP 323 and SER 289 respectively. |
Free Binding Energy (MM-GBSA)
The synthesized compounds and protein were prepared from the previously described methods and all the water molecules were deleted prior to the running of MM-GBSA prime. The free binding energy estimate was carried out using model Prime MMGBSA DG binds from Maestro 9.0 (Schrodinger Inc. USA). Table 2 is showing results of free binding energy concluding that the analogues M15, M17 and M8 likely disposed into the receptor PPAR-γ (1FM9). Glibenclamide chosen as standard drug was having binding energy of -48.58 Kcal/mol less than the most efficient ligands M15, M17 and M8 (-66.27, -66.21 and -66.21 Kcal/mol respectively). Binding energies of all synthetic entities are also shown in table 2 which are from -61.26 to-80.15 kcal/mol, resulting in more efficiency than standard.
 |
Table 2: Results of Molecular Docking and Free Binding Energy. |
Biological Evaluation
In vivo Oral glucose tolerance in normal rats
Derivatives M2, M4, M8, M9, M12, M13, M15, M17, M20 were separated through their glide scores in molecular modeling studies for testing oral glucose tolerance in usual rats. Results are shown in Table 3 and Fig. 4. The blood glucose level was increased from 172.05 and 165.85 mg/decilitre in control and glibenclamide dosed group respectively after giving glucose (at 30min) which shows hyperglycemic peak. Dispensing of derivatives increased glucose amount to a range of 161.75-173.98 mg/dL after thirty minutes of glucose consumption. After that, administration of novel drugs decreases the sugar level to small extend than induced after ninety minutes the normal group. At 90 min, blood glucose levels in novel derivatives reached glibenclamide-dosed level. Blood glucose reduction after giving glucose at ninety minutes was most compelling in M15, which is found to be even more potent than the standard glibenclamide-dosed group.
Table 3: OGTT results in Normal Rats
| Normal rats Group | Dose taken | 0min | 30min | 90min |
| Normal control | 0.1%CMC | 86.21±0.57 | 172.05±2.75 | 127.88±3.68** |
| Glibenclamide | 10mg/kg | 92.76±2.32 | 165.85±3.28 | 105.05±1.14** |
| M2 | 100mg/kg | 89.66±1.85 | 173.15±2.86 | 112.8±3.62** |
|
M4 |
100mg/kg | 86.90±2.62 | 169.23±4.28 | 109.13±1.50** |
|
M8 |
100mg/kg |
87.31±2.08 |
166.37±2.51 |
108.93±2.77** |
| M9 | 100mg/kg | 85.30±1.69 | 167.16±3.06 |
114.2±2.76** |
| M12 | 100mg/kg | 88.95±1.57 | 173.98±3.58 | 110.41±2.73** |
| M13 | 100mg/kg | 89.70±2.00 | 170.95±3.31 | 109.18±1.99** |
|
M15 |
100mg/kg | 88.95±1.93 | 166.98±3.93 |
104.38±1.60** |
| M17 | 100mg/kg | 87.68±1.89 | 161.75±2.85 | 107.88±2.41** |
| M20 | 100mg/kg | 87.96±3.10 | 170.40±4.06 | 111.93±2.28** |
Data represent (n=6) mean ± SEM, analyzed comparing to control through one-way ANOVA and Dunnett’s multiple comparison test , **p<0.01
 |
Figure 4: Graphical representation of in vivo Oral Glucose Tolerance Test. |
In vivo Antihyperglycemic evaluation in STZ induced diabetic rats
The selected synthesized fatty acid derivatives were dispensed to diabetic albino rats for regulating the glucose levels at different time intervals. Alterations were taken for a period of six hours to measure blood glucose level for each group as 0h, 2h, 4h, 6h after giving STZ (60mg/kg) intraperitoneally described in Table 4 and Fig. 5. The oral administration of glibenclamide (10mg/kg body weight) displayed notable decrease in sugar level in this period. Glibenclamide generated remarkable lowering of glucose amount to 103mg/decilitre in 6 h. During this duration there was very less reduction of glucose level in control group. Giving selected synthetic chemical entities (100 mg/kg body weight) presented drop in level of glucose in 6 h, slighter than produced by glibenclamide. M15 is found to be most potent even more than the standard as it reduced the blood glucose level by 104.24mg/dL in 6h which is more than standard glibenclamide (103mg/dL). Further we can conclude that M17 and M8 are also active when compared to standard. Compounds M2, M4, M9, M12, M13 and M20 are moderately active. Thus by the results of antidiabetic activity, it can be considered that further these fatty acid derivatives can be used as anti-hyperglycemic drugs with safer early onset of action.
Table 4: Antihyperglycemic activity results in diabetic rats
| Diabetic Rat Group | Dose taken | 0h | 2h | 4h | 6h |
| Diabetic control | 0.1%CMC | 223.23±2.36 | 219.48±2.36 | 216.16±2.301 | 207.16±2.14** |
| Glibenclamide | 10mg/kg | 217.16±2.51 | 198.13±1.99 | 169.66±1.53 | 113.7±2.65** |
| M2 | 100mg/kg | 223.75±2.29 | 192.81±2.26 | 163.58±2.89 | 125.51±1.95** |
| M4 | 100mg/kg | 221.78±2.87 | 192.81±2.48 | 163.26±2.37 | 123±2.28** |
| M8 | 100mg/kg | 224.45±1.19 | 192.5±1.71 | 158.3±2.99 | 121.66±3.22** |
| M9 | 100mg/kg | 222.38±1.18 | 201.33±1.573 | 166.76±4.75 | 127.71±2.50** |
| M12 | 100mg/kg | 221.2±1.58 | 197.95±1.43 | 175.05±3.52 | 124.5±2.05** |
| M13 | 100mg/kg | 223.03±3.86 | 201.38±3.59 | 163.58±4.78 | 124.35±2.29** |
| M15 | 100mg/kg | 217±1.44 | 194.7±3.34 | 159.56±2.87 | 112.76±1.57** |
| M17 | 100mg/kg | 223.75±2.29 | 192.81±2.26 | 163.58±2.89 | 120.76±2.98** |
| M20 | 100mg/kg | 219.91±3.86 | 196.45±3.47 | 165.3±2.73 | 123±2.28** |
Data represent (n=6) mean ± SEM, analyzed comparing to control through one-way ANOVA and Dunnett’s multiple comparison test, **p<0.01
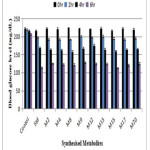 |
Figure 5: Graphical representation of in vivo antihyperglycemic results Click here to View figure |
SAR
Results of the synthesized novel compounds displayed that:
Either mono or disubstituted electron withdrawing groups, increases anti-hyperglycaemic activity as in M15 and M8 which contain ortho fluoro and orttho and para dichloro respectively.
Compound M17 was an exception in which electron donating group ie. methyl at ortho and para showed second highest antihyperglycaemic action.
Also the exsistence of electron donating groups on phenyl ring at 4th position results in moderate anti-hyperglycaemic potential as shown by compounds M13 and M20 which contain methyl and ethyl group respectively at para position.
Substituting phenyl moiety with some heterocyclic moiety moderately increases the anti-diabetic activity comparing to standard glibenclamide as in compound M12 having 2-methyl piperazine moiety.
Discussion
The focus in our work was on design, synthesize, perform docking and then investigating whether the selected synthesized derivatives of naturally occurring fatty acid could trigger reduction in glucose level in normal rats and anti-diabetic impact on diabetic rats. The conclusions indicate a possible utilization of these derivatives in diabetes as additive food. This present work although showed moderate hypoglycaemic potential but the synthesized products possess high anti-diabetic effect in STZ-induced albino rats. Diabetes mellitus is activated extensively by streptozotocin lethaly affecting pancreatic β-cells, accounting the secretion of insulin.25 Therefore diabetes activated by STZ is marked with steady increase in glucose level. As STZ initiateshyperglycaemia through demolishing of beta cells and by damaging function of kidney, glibenclamide displayed lenient hypoglycaemic activity in the rats.
At present diabetes strikes over and above 285 million population of world, anticipated to mount to 435 million by 2030 because of population rise, aging, deleterious diet, fleshiness, and desk bound job. The significant activities shown by compounds containing fatty acid and oxadiazole moiety are of different types like decreasing diabetes, antineoplastics, reducing inflammation and some other pharmacological activities which invoke them as vital analogue for evolution of novel remedial agents. These compounds arbitrate anti-diabetic effects through agonistic effects on PPAR gamma.
Fatty acids are chief nutritional component involved in the cardiovascular and metabolic diseases5. Foods containing saturated fatty acids are also more preferable for the type 2 diabetic patients.6 Recently it has been reported that substituted long chain fatty acids and its derivatives activate PPAR-γ and are their endogeneous ligands.9 PPAR-β and PPAR-γ are triggered by indistinguishable compounds like natural eicosanoids, fatty acids and other drugs.2,3 As fatty acids are significant in treating type II diabetes, this evoked us to plan this study having fatty acid integrated with oxadiazole heterocycle in search for the better anti-hyperglycaemic activities and safer profile than the available drugs.
Our results show that selected synthesized derivatives has anti-hyperglycaemic activity in accordance with surveys previously published. Although complete mechanism of action needs more exploration, we have proposed that fatty acid diet or the synthesized derivatives as drug can avert increasing glucose level in the STZ induced diabetic rats. The work proposed can conclude that the selected derivatives can decrease sugar level to normal by giving to longer time. This is the only work which shows that the derivative of natural occurring fatty acid having antidiabetic potential, this discovery allocate a basis for the use fatty acid derivatives having oxadiazole moiety for deterrence of diabetes and may help in prevention of type 2 diabetes mellitus. Certainly more research due to its indelible consequences in diabetic patients is desired.
Results of molecular docking demonstrated that compounds M15, M17 and M8 showed good docking score as -10.4, -10.2 and -10.0 respectively, well located in the active sites. The free binding energy lies between -80.2 to -61.3 kcal/mol which is significant as compared to that of standard (-48.6 kcal/mol). Compound M15 was identified as most active and can be used lead candidate with excellent anti-hyperglycaemic activity due to presence of electron withdrawing group ortho-fluoro increasing its anti-hyperglycaemic activity. M15 reduced the sugar elevation by 104.2 mg/decilitre which when compared to standard glibenclamide (103 mg/dL) is more. M8 also displayed good activity comparable to standard which was having electron withdrawing group chloro at ortho and para. M17 was an exception which carries electron donating group but showed good activity. All other synthesized derivatives showed moderate anti-hyperglycemic activity. Some derivatives showed low activity due to brief experimental time, so long period work may be required to get best results.
Therefore, considering that fatty acid derivatives possess a polar chain and an oxadiazole moiety at end which are required for good anti- hyperglycaemic activity, novel mannich base derivatives having this type of model were synthesized by substituted amines. All twenty two compounds showed good activity with compound M15 more potent than the standard drug due to the presence electron withdrawing group fluoro and thus can be further explored in search of potential new agents for type II diabetes. M17 and M8 could also act as lead to exploit its relevant therapeutic effect as anti-diabetic agent.
Conclusion
This research illustrates simple and efficient synthesis of novel mannich base derivatives of fatty acid showing significant and attractive anti-diabetic properties tested on wistar rats when compared with the standard, glibenclamide. Furthermore investigations can be done on these prominent candidates for the search of potent new agents for type 2 diabetes having bioactive core for PPAR agonists.
Acknowledgments
Ms. Garima Kapoor and Ms. Rubina Bhutani are indebted to DIPSAR, New Delhi, India for providing laboratory research facilities. We both are also thankful to DST (sanction order numbers DST/Inspire Fellow/2014/266 and DST/Inspire Fellow/2014/258 respectively), India for providing financial assistance in our research work.
References
- Murugan R, Anbazhagan S, Narayanan SS. Eur. J. Med. Chem. 2009, 44, 3272–3279.
CrossRef - Wolfrum C, Borrmann CM, Borchers T, Spener F. Proc. Natl. Acad. Sci. 2001, 98, 2323–2328.
CrossRef - Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Kobles CS, Devchand P, Wahli W, Willson TM, Lenhard JM, Lehmann JM. Proc. Natl. Acad. Sci. 1997, 94, 4318–4323.
CrossRef - Krey G, Braissant O, Horset FL, Kalkhoven E, Perroud M, Parker MG, Wahli W. Mol Endo. 1997, 11, 779-791.
CrossRef - Kotarsky K, Nilsson NE, Flodgren E, Owman C, Olde B. Biochem. Biophys. Res. Commun. 2003, 301, 406–410.
CrossRef - Schoonjans K, Staels B, Auwerx J. J. Lipid Res. 1996, 37, 907-925.
- Storm H, Thomsen C, Rasmussen EP, Christiansen C, Hermansen K. Diabetes care. 1997, 20 1807-1813.
CrossRef - Itoh Y, Kawamata Y, Harada M, Kobayashi M, Fujii R, Fukusumi S, Ogi K, Hosoya M, Tanaka Y, Uejima H, Tanaka H, Maruyama M, Satoh R, Okubo S, Kizawa H, Komatsu H, Matsumura F, Noguchi Y, Shinohara T, Hinuma S, Fujisawa Y, Fujino M. Nature. 2003, 22, 173-176.
CrossRef - Schopfer FJ, Cole MP, Groeger AL, Chen C, Khoo NKH, Woodcock SR, Bisello FG, Motanya UN, Li Y, Zhang J, Garcia-Barrio MT, Rudolph TK, Rudolph V, Bonacci G, Baker PRS, Xu HE, Batthyany CI, Chen YE, Hallis TM, Freeman BA. J. Biol. Chem. 2010, 285, 12321-12333.
CrossRef - Rauf A, Bandaya MR, Mattoo RH. Acta Chim. Slov. 2008, 55, 448–452.
- Hasan A, Thomas NF, Gapil S. Molecules. 2011, 16, 1297-1309.
CrossRef - Frank PV, Poojary MM, Damodara N, Chikkanna C. Acta Pharm. 2013, 63, 231–239.
CrossRef - Bhutani R, Pathak DP, Kapoor G, Husain A, Kant R, Iqbal MA. Bioorg. Chem. 2018, 77, 6–15.
CrossRef - Molinspiration software or free molecular property calculation services. Available from URL: www.molinspiration.com/cgi-bin/properties (last accessed 21.05.17).
- Ali MR, Kumar S, Afzal O, Shalmali N, Ali W, Sharma M, Bawa S. Arch. Pharm. Chem. Life Sci. 2017, 350, 1-13.
- Silva MM, Comin M, Duarte TS, Foglio MA, Carvalho JE, Vieira MC, Formagio ASN. Molecules. 2015, 20, 5360-5373.
CrossRef - Kumar NS, Pradeep T, Jani G, Silpa D, Kumar BV. J. Adv. Pharm. Tech. Res. 2012, 3, 57-61.
- Debnath B, Ganguly S. Asian J Pharm Clin Res. 2014, 7, 186-194.
- Siddiqui N, Alam MS, Ali R, Yar MS, Alam O. Med. Chem. Res. 2016, DOI 10.1007/s00044-016-1570-6.
CrossRef - Maestro. Version 9.0, Schrodinger, LLC, New York, NY.
- Avupati VR, Yejella RP, Akula A, Guntuku GS, Doddi BR, Vutla VR, Anagani SR, Adimulam LS, Vyricharla AK. Bioorg. Med. Chem. Lett. 2012, 22, 6442–6450.
CrossRef - Firke SD, Firake BM, Chaudhari RY, Patil VR. Asian J. Research Chem. 2009, 2, 57-161.
- Lee C, Lee HS, Cha YJ, Joo WH, Kang DO, Moon JY. Prev. Nutr. Food Sci. 2013, 18, 169-174.
CrossRef - Jain K, Malviya S, Gupta AK, Kharia A. IJPSR. 2014, 5, 5025-5041.
- Nazreen S, Alam MS, Hamid H, Yar MS, Shafi S, Dhulap A, Alam P, Pasha MAQ, Bano S, Alam MM, Haider S, Ali Y, Kharbanda C, Pillai KK. Eur. J. Med. Chem. 2014, 87, 175-185.
CrossRef - Al-Abdullah ES, Al-Tuwaijri HM, Hassan HM, Haiba ME, Habiband EE, El-Emam AA. Int. J. Mol. Sci. 2014, 15, 22995-23010.
CrossRef - Ali Z, Akhtar MJ, Siddiqui AA, Khan AA, Haiderand MR, Yar MS. Arch. Pharm. Chem. Life Sci. 2017, 349, 1-11.
- Kumar RBS, Kar B, Dolai N, Bala A, Haldar PK. Asian Pac J Trop Dis. 2012, 139-143.

This work is licensed under a Creative Commons Attribution 4.0 International License.









