An Eco-friendly Approach for Synthesis of Platinum Nanoparticles using Leaf Extracts of Jatropa Gossypifolia and Jatropa Glandulifera and its Antibacterial Activity
U. Jeyapaul1 , Mary Jelastin Kala2, A. John Bosco1, Prakash Piruthiviraj3 and M. Easuraja4
, Mary Jelastin Kala2, A. John Bosco1, Prakash Piruthiviraj3 and M. Easuraja4
1,2PG and Research Department of Chemistry , St. Xavier's College (Autonomous), Palayamkottai, 627002 Affiliated to Manonma
1Department of Chemistry, SRM Institute of Science and Technology, Faculty of Engineering and Technology, Kattankulathur, Kancheepuram-603203, Tamilnadu, India.
3DRDO – BU Center for Life Sciences, Bharathiyar University Campus, Coimbatore, Tamil Nadu, India.
4Department of Chemistry, Arul Aanadar College (Autonomous), Karumathur, Tamilnadu, India.
*Corresponding Author E-mail: jpaulser83@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/340223
Article Received on : November 26, 2017
Article Accepted on : December 30, 2017
In present scenario, ecofriendly approach for the synthesis of noble metals nanoparticles by means of various plant extracts has became a great interest in the field of nanotechnology. This green route schemes are eco-friendly, simple, less toxic and environmental benign. In this paper, an environmental friendly approach is presented for the formation of Platinum nanoparticles using aqueous leaf extracts of Indian medicinal herbs such as Jatropa Gossypifolia and Jatropa Glandulifera as an efficient reducing and capping agents. A great transformation pathway of platinum ions to platinum nanoparticles was obtained by employing the Jatropa Gossypifolia and Jatropa Glandulifera leaves broth with a reaction temperature of 100°C. Biosynthesized Platinum Nanoparticles were successfully characterized by Ultraviolet-Visible spectroscopy. FTIR studies confirmed the presence of amine, amide. The -SO2 functional group were found to be prone for reduction of platinum metal to platinum nanoparticles. Field Emission scanning electron microscope (SEM) gives information about the shape and size of the formed nanoparticles. Average Platinum Nanoparticles particle size distribution ranges from nanometer to micrometer. Transmission Electron microscope predominantly indicates the biosynthesized Platinum Nanoparticles that are dodecahedron and some kind of cubic in nature. In addition, biologically synthesized Platinum Nanoparticles of leaf extracts exhibits a immense potent of antibacterial activity against pathogenic bacteria's.
KEYWORDS:Jatropa Gossypifolia; Platinum Nanoparticles; FT-IR; FE-SEM; Antibacterial Activity
Download this article as:| Copy the following to cite this article: Jeyapaul U, Kala M. J, Bosco A. J, Piruthiviraj P, Easuraja M. An Eco-friendly Approach for Synthesis of Platinum Nanoparticles using Leaf Extracts of Jatropa Gossypifolia and Jatropa Glandulifera and its Antibacterial Activity. Orient J Chem 2018;34(2). |
| Copy the following to cite this URL: Jeyapaul U, Kala M. J, Bosco A. J, Piruthiviraj P, Easuraja M. An Eco-friendly Approach for Synthesis of Platinum Nanoparticles using Leaf Extracts of Jatropa Gossypifolia and Jatropa Glandulifera and its Antibacterial Activity. Orient J Chem 2018;34(2). Available from: http://www.orientjchem.org/?p=44038 |
Introduction
Nanotechnology is an upcoming research field having immense advantages in science and technology for the purpose of industrialized new materials at the nanoscale level. At present, biosynthetic methods employing either microorganisms or plants extract has emerged as an alternative to more complex chemical synthetic procedures to obtain nanomaterials. Particularly, Bio synthesis of nanoparticles (NPs) has upsurge in the field of nanobiotechnology to make novel materials that are ecologically approachable, cost effectual, stable NPs with great importance for wider applications especially in the field of medicine.1. In today’s modern world, interesting features of nonmaterial’s have become popular due to their broad spectrum of application in biological and medical fields especially in wound healings2. Predominantly, Platinum based alloys focusing a greater attention towards a various types of catalytic applications such as degradation of organic dyes 3, oxidation and reduction reactions 4-5 and catalytic hydrogenation conversions 6. Subsequently, precise and controlled biofabricated Platinum Nanoparticles shape and size have been demonstrated through various physical and chemical methods. However, the above mentioned methods were followed in stringent conditions such as toxic chemicals and harmful solvents7-8. To overcome this strategy, biogenic synthesis based on plant extracts was found to be the best choice for producing stable metal nanoparticles. Young plants, plant extracts, leaf broths and plant biomass are received more attention to the researchers due to the fact of low cost, less toxic, highly effective and environmental benevolent candidates for the extracellular green synthesis of metal NPs. During the last few decades, few efforts have been made towards the intracellular or extracellular biosynthesis of Platinum Nanoparticles using microbes under mild conditions which is devoid of additional capping agents9–12. However, For instance, C.Soundarrajan and co-workers 13 have reported the green synthesis of Platinum nanoparticles using Ocimum sanctum leaf extract. In 2010 J.Y Song has reported the extracellular production of Platinum nanoparticles using Diopyros kaki leaf extract 14. Additionally, numerous plant extracts mediated Platinum Nanoparticles, consisting medicinal herbs of alfalfa 15 , Cinnamomum camphora leaves 16 , Punica granatum 17 Herbal leaves of Bidens Tripartitus18 Cochlospermum gossypium 19, are reported. Exceptionally medicinal values of neem leaves 20, Aloe- vera leaf extracts 21 , tea and Camellia sinensis 22-23 extracts have also been employed to synthesis core shell metallic Au, Ag and Pt nanoparticles. Recently, green synthesis of Platinum Nanoparticles by using Fumaria officinalis L plant extracts 24 has also been carried out. A detailed survey of literature reveals that there is no report on the biosynthesis of Platinum Nanoparticles using aqueous leaf extracts of Jatropa Gossypifolia and Jatropa Glandulifera till now.
Jatropa Gossypifolia and Jatropa Glandulifera are an indian medicinal leaves belonging to euphorbiaceae family. Mostly, this types of genus plants are grown in Southern part of India, particularly, in Sirumali Dindigul district, Tamilnadu, India. Moreover, this medicinal valuable leaves having enormous number of anticancer and antibacterial medicinal values. This present study aims at developing the biosynthesis of Platinum Nanoparticles using the aqueous extract of Jatropa Gossypifolia and Jatropa Glandulifera. The synthesized Platinum Nanoparticles were characterized by UV-Visible spectroscopy, Field Emission- Scanning electron microscopy (FESEM) , High Resolution Tunneling electron microscopy (HR-TEM), X-ray diffraction studies (XRD), and Fourier transform-Infra red spectroscopy (FT-IR) . At last the antibacterial activity for synthesized Platinum Nanoparticles were examined by Well -disc diffusion method.
Materials and Methods
Chemicals and Reagents
Hexachloro Platinic acid, (H2PtCl6 .6H2O) 99.9% purity were procured from Sigma -Aldrich Mumbai, India. Fresh green and mature leaves of Jatropa Gossypifolia and Jatropa Glandulifera plants were collected in March 2017 from Dindigul District, Tamilnadu, India. Double-sterilized Milli-Q water was used throughout the experiments. Unless otherwise stated, the other chemicals and solvents were used under analytical grade without further purification.
 |
Figure 1: Digital specimen of Leaf Jatropa Gossypifolia |
 |
Figure 2: Digital specimen of Leaf Jatropa Glandulifera |
Preparation of Leaf Extract
Fresh leaves of Jatropa Gossypifolia and Jatropa Glandulifera were collected in the month of march near to sirumali hills Dindigul District. These herbs were taxonomically identified and authenticated by Head of the Department of Botany, St. Xavier’s College Palayamkottai, Tamilnadu, India. Healthy and young leaves of plants portion were selected and washed under tap water to remove the sand particles which is tightly adhere over to the leaves surface and then rinsed with Millipore -Milli-Q water. The Fresh and steriled leaves were completely dried at room temperature. The dried leaves were milled well and collected in an air- tight container bottle for further use. The aqueous leaf extracts of Jatropa Gossypifolia and Jatropa Glandulifera were prepared as follows. About 30 g dried leaves were dissolved in 100 mL of de-ionized water in a 250 mL of Erlenmeyer flask and the above contents were boiled for 30 minutes and the formed products was subjected to deep freezing. Suspensions of these extract was filtered through Whatman NO.1 filter paper. The obtained clear leaf extracts were used for the synthesis of Platinum nanoparticles.
Synthesis of Platinum Nanoparticles
In order to synthesis the Platinum nanoparticles, we followed the supporting literature of the protocol described by Preeti Dauthal et al 25 was followed with a little modifications. 100 ml of 1mM of metal solution of H2PtCl6 was prepared. From the above prepared metal solution 90 ml of 1mM H2PtCl6.6H2O were taken and then it was stirred continuously for 1 hour at 30oC. To the above solution, about 10 ml of prepared plant extract was added dropwise in the ratio of 1:9. Again, the mixture was stirred for more than 20 minutes. The color change (dark brown color) indicated the bio reduction of the platinum metals. The Bio-reduced component was confirmed by using UV-visible spectrophotometer.
Characterisation of Platinum Nanoparticles
Biosynthesised Platinum nanoparticles were analysed by means of UV-Visible (UV-VIS) spectrophotometer (SHIMADZU model UV-1650 PC, Japan) at wavelength of 200-800 nm. The dried powder of Indian leaves were analysed by X-ray diffractometer using PAN analytical X’pert-PRO diffractometer with CuKα radiation in the range of 200 and 800 2q values with a scanning rate of 4o/min. FTIR are carried out for the biogenic green samples with PERKIN ELMER FT-IR 4100 instrument. in transmission mode at a resolution 4 cm-1 in KBr pellets. The pelletized KBr containing dried leaf extract powder was used as control level. Size and morphology of the samples were analysed by Field-Emission Scanning electron microscope (FESEM) and Transmission Electron Microscope (TEM). To predict the shape of the morphology SEM were performed by using (FE-SEM, Model S-4200 Hitachi, Japan). TEM analyses were examined by placing a drop of green synthesised Platinum Nanoparticles on carbon-coated grids and allowed the system get dried completely in a vaccum oven and then the instruments were operated at 200 kV.
Cyclic Voltammetry
Redox prototype for the biogenic green samples were carried out in an electrochemical workstation (CH-SP300 Biologic France) using a conventional three-electrode system at room temperature. Green synthesized samples of PlatinumNanoparticles were used as the working electrode, an Ag/AgCl as reference electrode and a platinum foil as the counter electrode along with 1.0 M KCl (99% purity, Sigma Aldrich, Mumbai) served as electrolyte solution.
Antibacterial Studies
The antibacterial activity of silver nanoparticles was explored utilizing discs diffusion assay 26. Good biosynthesised Platinum Nanoparticles using the Jatropa leaf extract were tested for the antibacterial activity by standard disc diffusion method. Gram-negative bacteria like Escherichia coli, Klebsiella pneumonia, Pseudomonas aeruginosa and Gram-positive Bacillus licheniformis, Staphylococcus epidermidis, Staphylococcus aureus were employed as bacteria strains for the antibacterial studies. This Wholesome culture was again sub-cultured in nutrient broth and the strain was evenly spread on sterilized Mueller Hinton Agar (MHA) petriplates. About 4 mm diameters of sterile circular discs (HIMEDIA – SD067) were performed on the MHA plates and the weight percentage of each loaded nanoparticles were placed over the appropriate disc at 10 mcg/disk. Tetracycline 10 mcg were chosen as a model antibiotic substrate were well thought-out as a control. The treated plates were incubated for 24 hours at 37°C in a sterile condition and dark region. Triplicate plates were maintained for each organism. After incubation, the percentage of inhibition (mm) was calculated.
Results and Discussion
UV-Visible Absorption Spectral Studies
Biosynthesised leaf extracts of Platinum Nanoparticles were determined by UV-Visible absorption spectral measurements. Upon the addition of H2PtCl6 .6H2O to the leaf extracts of Jatropa Gossypifolia and Jatropa Glandulifera, we observed a sudden change of color from light to dark brown within 30 minutes at room temperature. It indicates the formation of Platinum Nanoparticles. Moreover, being a strong Ligand to Metal charge transfer process between Pt 4+ and Cl – ions, the absorption spectrum of Platinum Nanoparticles showed a peak value at 260 nm, which strongly indicated the formation of platinum nanoparticles 27 as shown in Fig. 3. Due to the good interfering effect between reaction time and the surface Plasmon resonance the sharp bands exhibits a red shift value at 260 nm which confirmed the formation of small spherical Platinum Nanoparticles. Similarly it was also noticed a strong absorption peak at 260 nm is attributed to Jatropa Glandulifera leaf extract. Suddenly the peak value at 260 nm decreases and reinstated by wide absorption spectrum which steadily increases in intensity from visible to the ultraviolet region, confirmed that Pt4+ ions were completely reduced to Pt0 28. Fig. 3 and Fig. 4 represent the UV-Visible spectrum of PlatinumNanoparticles synthesised by a green approach.
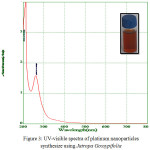 |
Figure 3: UV-visible spectra of platinum nanoparticles synthesize using Jatropa Gossypifolia |
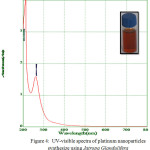 |
Figure 4: UV-visible spectra of platinum nanoparticles synthesize using Jatropa Glandulifera |
FT-IR Analysis
Fourier Transform Infrared Spectroscopy was found to be an important technique to identify th. In FTIR spectrum the major absorption bands for J. Gossypifolia and Glandulifera at 3542cm-1, 2058 cm-1,1637cm-1,1354 cm-1 , 3451 cm-1, 2046 cm-1, 1353 cm-1 is due to -OH bending and -NH bending vibrations as shown in Fig.5. Strong peak at 3542 cm-1 was due to the presence of -OH groups present in the leaf extracts. 2046 cm-1 peak value were confirmed the presence of -NH stretching vibrations which is the major candidate for reducing the metal ions. Typically the most intense band at 1353 cm-1 was due to the presence of -CH3 symmetrical deformations in the compounds. The same similar FT-IR spectrum was obtained for Jatropha Glandulifera as shown in Fig.5. The peak value at 3452 cm-1 confirmed -OH bending vibrations. Strong intense peak value at 1637 cm-1 may be due to the presence of -NH bending vibrations in the leaf extracts. In addition, the peaks due to sulphur such as S-H, S-S, and C-S bonds, were shown by weak infrared peaks. Compounds, such as sulfides containing C-S and S-S bonds, show stretching frequency value at 695 cm−1 and 607 cm−1 respectively.
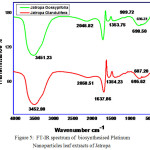 |
Figure 5: FT-IR spectrum of biosynthesised Platinum Nanoparticles leaf extracts of Jatropa |
Gossypifolia and Jatropa Glandulifera
X-ray Diffraction Studies (XRD)
The crystalline nature of the platinum nanoparticles was confirmed by X-ray Diffraction analysis. XRD pattern clearly predicted that Platinum Nanoparticles were formed by reduction of leaf extracts of Jatropha Gossypifolia and Jatropha Glandulifera in Fig.6. The characteristic diffraction peaks at 2ϴ values were predicted at 39º, 46º, 67º,81º and 85º which corresponds to the (111), (002), (022), (113) and (222) respectively. According to JCPDS Card No 98-006-0819 , XRD pattern revealed the formation of Platinum Nanoparticles structure as cubic in nature.
 |
Figure 6: XRD spectrum of biosynthesised Platinum Nanoparticles leaf extracts of Jatropa |
Gossypifolia and Jatropa Glandulifera
Field-Emission Scanning Electron Microscope
FESEM reveals the surface morphology of the biosynthesised Platinum Nanoparticles. The surface morphology and the particle size were observed by SEM. The formed Platinum Nanoparticles were examined from nanometer level to micrometer level. It was observed that Jatropha Gossypifolia approximately appears as a rectangular, cubic and hexagonal like structure while Jatropha Glandulifera appears as irregular spherical plates. The FESEM micrographs also reveals the crystalline particles which are slightly agglomerated, This may be due to the excess adsorption of water molecules captured onto the metal particles. Fig.7 and Fig.8 shows the FESEM images of biosynthesised Platinum Nanoparticles of aqueous leaf extracts of J. Gossypifolia and J. Glandulifera
 |
Figure 7: FESEM images of Platinum Nanoparticles of aqueous leaf extracts Jatropha Gossypifolia |
 |
Figure 8: FESEM images of Platinum Nanoparticles of aqueous leaf extracts of Jatropha Glandulifera |
Transmission Electron Microscopy (TEM) Analysis
The TEM micrograph of Platinum Nanoparticles of green synthesised leaf extracts are shown in Fig. 9 and Fig.10 respectively. TEM revealed the formation of spherical and dodecahedron like structure inset in (Picture 9) formed in the mean particle range of 20 nm. Typically, a similar TEM image of Jatropha Glandulifera appears in the average particle size of 100 nm. From the micrograph it was clearly observed that the prepared Platinum Nanoparticles are relatively spherical, dodecahedron and small cubic in nature with some deviations respectively.
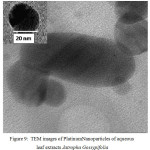 |
Figure 9: TEM images of Platinum Nanoparticles of aqueous leaf extracts Jatropha Gossypifolia |
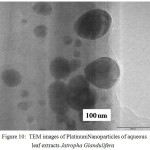 |
Figure 10: TEM images of Platinum Nanoparticles of aqueous leaf extracts Jatropha Glandulifera |
Cyclic Voltammetry
To explore the main advantages of silver nanoparticles of aqueous leaf extracts for redox process , we studied their electrochemical performance by cyclic voltammetry. All the experiments were performed with three electrode configuration cell-set up. Platinum foil and a saturated calomel electrode (SCE) were used as the counter and reference electrodes respectively. 1 mol L-1 KOH solution was used as the working electrolyte. Fig. 11 and Fig. 12 show CV curves of Jatropa Gossypifolia and Jatropa Glandulifera at a scan rate of 10 mV s-1. At a scan rate of 0.05 mV s-1, cyclic voltammetry curves exhibits a major oxidation or anodic peak at -0.5 mV s-1 and reduction or cathodic peak at 1.0 V (s Ag/AgCl). Anodic and cathodic current were strongly increase the oxidized and reduced peak potential with increase in scan rate. From these observations it was identified that oxidation peak moved towards on the a little positive shift, while reduction peak is gradually shifted in negative potential.
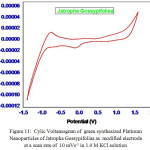 |
Figure 11: Cylic Voltamogram of green synthesized Platinum |
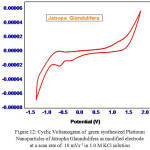 |
Figure 12: Cyclic Voltamogram of green synthesized Platinum |
Antibacterial Studies
After 24 hours of incubation, the zone of inhibition was calculated in all plates (width in mm). For six different bacterial strains both gram positive and negative, the development zone of inhibition (Table.1) was observed for the concentration of 10 mcg per discs of synthesized Plainum Nanoparticles. For gram negative bacterial against activity of PG and PB mean value of zone of inhibition (mm) was 5.0 and 6.0. For PG and PB against activity of gram positive bacterial mean value of zone of inhibition (mm) was 3.3 and 3.3. In contrast antimicrobial activity of nano particles is opposite to that of Gram-positive and Gram-negative bacteria was basically due to the strong electrostatic interface between nanoparticles and bacteria. Most of the nanoparticles exhibited enhanced antimicrobial behavior against Gram-negative bacteria. It is primarly because of difference in the structure and cell wall composition. The Prime difference between Gram-negative and Gram-positive bacteria is the existence of external membrane and the periplasmic space, that is adjacent to the external membrane. The outer membrane of Gram-negative bacteria is lipid bilayer, whereas the inner membrane composed of phospholipids. The cell wall of Gram-negative bacteria has peptidoglycan layer (7–8 nm) which gives hardness to cell wall. Nanoparticles act together with cell wall of bacteria by the electrostatic attraction. Due to thin peptidoglycan layer, nanoparticles might enter more easily, and therefore, antimicrobial activity was found to be unsuitable for Gram-negative bacteria 29. Due to the presence of rigid cell wall of Gram-positive bacteria, the antimicrobial activity of Gram-negative bacteria was higher as compared to Gram-positive bacteria.
 |
Figure 13 : Zone incubation (mm) activity of platinum Jatropa Gossypifolia and platinum Jatropa Glandulifera against pathogenic bacterias |
Table 1: Antimicrobials activity of synthesised platinum nanoparticles
| S.No | Bacterial strain | Class |
Mean values of zone of incubation in mm |
|||
| Tetracycline | PG | 10 mcg/disk | PB | |||
| 1. | Bacillus licheniformis | Gram Positive |
16 |
0 |
– |
|
| 2. | Staphylococcus epidermidis | Gram Positive |
20 |
0 |
– |
|
| 3. | Staphylococcus aureus | Gram Positive |
09 |
10 |
10 |
|
| 4. | Escherichia coli | GramNegative |
14 |
0 |
0 |
|
| 5. | Klebsiella pneumoniae | GramNegative |
13 |
08 |
09 |
|
| 6. | Pseudomonas aeruginosa | GramNegative |
20 |
07 |
09 |
|
PG – Platinum Jatropa Gossypifolia ; PB- Platinum Jatropa Glandulifera;
Conclusion
Green route has been established for the synthesis the Platinum Nanoparticles using Jatropha Gossypifoliaa and Jatropha Glaundulifera leaf extracts. The formation of Platinum Nanoparticles was successfully confirmed by UV-Visible spectrometery. Dodecahedron shaped and small cubic like structure were identified in TEM analysis within the range of 100-200 nm. XRD reveals that the nature of formed Platinum Nanoparticles has been crystalline in nature. FT-IR strongly confirms the presence of amine groups, carboxyl and carbonyl groups which plays a major role in the reduction of Pt 4+ to Pt0 ions. Furthermore, the prepared Platinum Nanoparticles has shown a good antibacterial activity against gram positive and gram negative pathogenic bacteria’s. So the green route synthesised Platinum Nanoparticles finds a wide application in the environmental concern and also controls the growth of the bacteria.
Acknowledgments
The authors of this present manuscript, Mr. U. Jeyapaul and Dr. A. John Bosco are equally contributed in this work. The above authors are very much grateful to the Department of Chemistry St.Xavier’s College, Palaymkottai, Tamilnadu India for providing the research facilities to carry out this work successful. Authors are sincerely thank to SRM Nanotechnology Research Institute for providing the facilities for SEM and XRD.
References
- Kim, J.S.; Kuk, E.; Yu, K.N.; Kim, J.H.; Park, S.J.; Lee, H.J.; Kim, S.H.; Park, Y.K.; Hwang, C.Y.; Kim, Y.K.; Lee, Y.S.; Jeong, D.H.; Cho, M.H.; Nanomedicine. 2007, 3, 95.
CrossRef - Franck, F.; Morley, K. S.; Ben, W.; Sharp, B. L.; Arnold, P. L.; Howdle, S. M.; Roger, B.; Brown, P. D.; Winship, P. D.; Reid, H. J.; J. Antimicrob. Chemother. 2004 , 54, 1019.
CrossRef - Venu, R.; Ramulu, T.; Anandakumar, S.; Rani, V.; Kim, C.; Colloids Surf. A 2011, 384, 733.
CrossRef - Wang, S.; Jiang, S.P.; Wang, X.; Guo, J.; Electrochim. Acta 2011, 56, 1563.
CrossRef - Esumi, K.; Isono, R.; Yoshimura, T.; Langmuir 2004, 20, 237.
CrossRef - Cheng, H.; Xi, C.; Meng, X.; Hao, Y.; Yu, Y.; Zhao, F.; J. Colloid Interf. Sci. 2009, 336, 675.
CrossRef - Mukherjee, P.; Roy, M.; Mandal, B.P.; Dey, G.K.; Mukherjee, P.K.; Ghatak, J.; Tyagi, A.K.; Kale, S.P.; Nanotechnology 2008, 19, 075103.
CrossRef - Kwi Jong, L.; Byung Ho, J.; Junrak, C.; Young, L; Jaewoo, J.; Yong Soo,O.; Nanotechnology 2007 , 18, 335601.
CrossRef - Riddin, T.L.; Gericke, M.; Whiteley, C.G.; Nanotechnology 2006, 17, 3482.
CrossRef - Konishi,Y.; Ohno, K.; Saitoh,N.; Nomura,T.; Nagamine, S.; Hishida, H.; Y. Takahashi, Y.; Uruga, T.; J. Biotechnol. 2007, 128, 648.
CrossRef - Riddin, T.; Gericke, M.; Whiteley, C.G.; Enzyme Microb. Technol. 2010, 46, 501.
CrossRef - Govender,Y.; Riddin, T.L.; Gericke, M.; Whiteley, C.G.; J. Nanopart. Res. 2010 12, 261.
CrossRef - Soundarrajan, C.; Sankari, A.; Dhandapani, P.; Maruthamuthu, S.; Ravichandran, S.; Sozhan, G.; Palaniswamy, N.; Bioprocess Biosyst. Eng. 2012, 35, 827.
CrossRef - Song, J.Y.; Kwon, E.-Y.; Kim, B.S.; Bioprocess Biosyst. Eng. 2010, 33, 159.
CrossRef - Gardea-Torresdey, J.L.; Gomez, E.; Peralta-Videa, J.R.; Parsons, J.G.; Troiani, H.; Jose, M.; Yacaman, Langmuir 2003, 19, 1357.
CrossRef - Huang, J.L.; Lin, L.Q.; Li, Q.B.; Sun, D.H.; Wang, Y.P.; Lu, Y. H.; He, N.; Yang,K.; Yang,X.; Wang, H.X.; Wang, W.T.; Lin, W.S.; Ind. Eng. Chem. Res. 2008, 47,6081.
CrossRef - Dauthal, P.; Mukhopadhyay, M.; J. Ind. Eng. Chem. 2015, 22, 185.
CrossRef - Dobrucka,R.; J. Inorg Organomet Polym. 2015, 11, 1.
- Selvi, K.; Vinod, V.T.P.; Tanuja,K.; Raju, D.; Karuna, R.; J. Environ. Radioact. 2015, 33, 148.
- Shankar, S.S.; Rai, A.; Ahmad, A.; Sastry, M.; J. Colloid Interface Sci. 2004, 275, 496.
CrossRef - Chandran, S.P.; Chaudhary, M.; Pasricha, R.; Ahmad, A.; Sastry, M.; Biotechnol. Prog. 2006, 22, 577.
CrossRef - Nadagouda, M.N.; Varma, R.S.; Green Chem. 2008, 10, 859.
CrossRef - Vilchis-Nestor,A.R.; Sánchez-Mendieta,V.; Camacho-López M.A.; GómezEspinosa, R.M.; Camacho-López, M.A.; Arenas-Alatorre, J.A.; Mater. Lett. 2008, 62 , 3103.
CrossRef - Dobrucka, R.; Saudi J. Biolo. Sci. 2016, 11, 012.
- Preeti, D.; Mausumi, M.; J. Ind. Eng. Chem. 2015, 22, 185.
CrossRef - K. Mallikarjuna, N. J. Sushma, B.V. S.Reddy, G. Narasimha, B. D.P. Raju, Int. J. Chem. Anal. Sci. 4 (2013) 14-18
CrossRef - Zhao, S.Y.; Chen, S.H.; Wang, S.Y.; Li, D.G.; Ma, H.Y.; Langmuir. 2002, 18, 3315.
CrossRef - Furlong, D.N.; Launikonis, A.; Sasse, W.H.; Sanders, J.V.; J. Chem. Soc. Faraday Trans. 1984, 80, 571.
CrossRef - Pandit, Raksha, Mahendra Rai, and Carolina A. Santos, Environ . Chem. Lett 15, 443-452
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









