Allylxanthone Derivatives as Xanthine Oxidase Inhibitors: Synthesis, Biological Evaluation and Molecular Docking Study
Thongchai Khammee1 , Warakorn Jongsu1, Mayuso Kuno2 and Sunit Suksamrarn2
, Warakorn Jongsu1, Mayuso Kuno2 and Sunit Suksamrarn2
1Department of Chemistry, Faculty Science and Technology, Phranakhon Rajabhat University, Bangkok, 10220, Thailand.
2Department of Chemistry, Faculty Science, Srinakharinwirot University, Bangkok, 10110, Thailand.
Corresponding Author E-mail: thongchai.k@pnru.ac.th
DOI : http://dx.doi.org/10.13005/ojc/340104
Gout is one of the most severe health problems of the aged and is caused by high levels of uric acid in the blood. The inhibition of Xanthine oxidase (XO) is one strategy to retain gout disease. Oxygenated xanthones and derivatives have been shown many important biological activities. However, some xanthones have the small amount of nature and its sulfur analogs, thioxanthone has not been well studied in their bioactivity. A series of hydroxyxanthones and allylxanthones analogous 3-5 have been synthesized and screened for their anti-XO activity. Leads to the discovery of 2,4-diallyl-1,3-dihydroxythioxanthone (5b) as the most active inhibitor with IC50 = 0.69±0.02 mM. Consequent molecular docking analysis by AutoDock 4.2 indicated that the most active compound (5b) inhibits XO by accommodated at the binding site of XO.
KEYWORDS:Xanthine Oxidase Inhibitors; Xanthone; Thioxanthone; Molecular Docking
Download this article as:| Copy the following to cite this article: Khammee T, Jongsu W, Kuno M, Suksamrarn S. Allylxanthone Derivatives as Xanthine Oxidase Inhibitors: Synthesis, Biological Evaluation and Molecular Docking Study. Orient J Chem 2018;34(1). |
| Copy the following to cite this URL: Khammee T, Jongsu W, Kuno M, Suksamrarn S. Allylxanthone Derivatives as Xanthine Oxidase Inhibitors: Synthesis, Biological Evaluation and Molecular Docking Study. Orient J Chem 2018;34(1). Available from: http://www.orientjchem.org/?p=42681 |
Introduction
Xanthine oxidase (XO) is a vital enzyme that catalyzes the oxidation of hypoxanthine to xanthine and the oxidation of xanthine to uric acid in the purine pathway and plays a significant role in hyperuricemia and gout1. There are two conventional treatments for gout that are used anti-inflammatory agents to relieve the symptoms of the disease and XO inhibitors. XO inhibitors are utilized to treat gout by blocked the synthesis of uric acid. Among of XO inhibitors, allopurinol is a standard drug to treat gout and acts as XO inhibitor. The use of allopurinol is restricted by adverse side effects including hypersensitivity reaction, renal toxicity2, Steven-Johnson syndrome and epidermal necrolysis3. Therefore, the search for new pharmacophores for XO inhibitor remains to continue. Phenolic-containing compounds are one of the largest classes widely found in natural sources and have been described to hold promise for human health benefits4. Hydroxyxanthones are a class of naturally occurring polyphenols that have shown to possess a broad range of biological activities5, including their XO inhibitory activity6. However, its sulfur analogs, thioxanthone, have never been reported. The bioactive thioxanthone7 and some xanthones are found in a limited amount in nature. Our interest in the bioactive compound along with their safety concern8 prompted us to explore the new anti-XO substances. In this report, we have synthesized a series of hydroxyxanthones and hydroxythioxanthones and screened for their anti-XO activity. The compound with the highest activity was then selected for molecular docking analysis to observe its preferred binding orientation to the enzyme target.
Material and Methods
General Methods
1H- and 13C-NMR spectra were recorded on a Bruker Avance 300 FT-NMR spectrometer, operating at 300 MHz (1H) and 75 MHz (13C). The NMR chemical shifts were referenced to the residual solvent peaks (dH 7.24 and dC 77.00 for CDCl3, dH 2.04 and dC 29.8 for acetone-d6). ESI-HR-TOF-MS analyses were conducted using a Bruker maXis™ 4G mass spectrometer. All flash column chromatography (CC) was performed on Merck silica gel 60 (particle size of 230-400 mesh). All reagents were purchased from commercial sources and used without further purification. The reactions progression were monitored by TLC which was operated on precoated silica gel GF254 preparative TLC plates. The spots on TLC were detected under UV light and by spraying with anisaldehyde-H2SO4 reagent followed by heating. Melting points were examined with Griffin melting point apparatus, and are uncorrected.
Preparation of 1,3-Dihydroxyxanthone (3)
Methanesulfonic acid (36.5 g, 25 mL, 0.4 mol) was added in a round-bottom flask (250 mL). Phosphorus pentoxide (P2O5, 3.6 g, 25.4 mmol) was added in one portion which was dissolved in 1-2 hr9. Then the solution was added to a mixture of salicylic acid (1, 1.38 g, 10 mmol), phloroglucinol (2, 1.26 g. 10.0 mmol) and heating at 80 °C for 30 min. The progression of the reaction was monitored by TLC. After completion of the reaction, the mixture was poured into ice-water to give the orange solid. The obtained solid was collected by filtration, and washed with distilled water, dried in air. The orange residue was then purified by flash CC eluting with 10% acetone-hexane obtained a yellow solid of the desired products (3) in 71.0 % yield10. 1,3-Dihydroxyxanthone (3), Yield: 71%; m.p.: 254-255°C, lit. 255-256°C 10, Rf: 0.50 (30% acetone-hexane), 1H-NMR [acetone-d6] δ: 6.25 (d, J = 2.0 Hz, 1H, H-2), 6.41 (d, J = 2.0 Hz, 1H, H-4), 7.43 (ddd, J = 8.6, 8.1, 0.9 Hz, 1H, H-7), 7.50 (dd, J = 7.1, 0.9 Hz, 1H, H-5), 7.81 (ddd, J = 8.6, 7.1, 1.6 Hz, 1H, H-6), 8.16 (dd, J = 8.1, 1.6 Hz, 1H, H-8), 12.89 (s, 1H, 1-OH), HR-TOF-MS (ESI, +ve) m/z: 229.0492 [M+H]+ (calcd 229.0495 for C13H8O3S +H).
Preparation of 1,3-dihydroxythioxanthone (5)
A mixture of MsOH (14.6 g, 10 mL, 0.1 mol) and alumina (Al2O3 acidic type, 0.3 g, 2.9 mmol) was added into thiosalicylic acid (4, 1.34 g, 0.09 mmol) and then phloroglucinol (2, 1.26 g, 10.0 mmol). The mixture was heated in an oil bath at 110 °C with stirring for 20 min. The reaction progress was monitored by TLC. After completion of the reaction, the mixture was poured into water and extracted with EtOAc (3 × 20 mL). The EtOAc layer was washed with a saturated aq. NaHCO3 (50 mL). The organic layer was dried over Na2SO4 and evaporated in vacuo to give a red residue. Chromatography of the crude product was performed on a column of silica gel eluted with 5% acetone-hexane to give a yellow solid of desired compounds (5) in 65% yield11. 1,3-dihydroxythioxanthone (5); m.p.: 228-230°C, lit. 232–233°C11, Rf: 0.35 (30% acetone-hexane), 1H NMR [Acetone-d6] δ: 6.36 (d, J = 2.2 Hz, 1H, H-2), 6.64 (d, J = 2.2 Hz, 1H, H-4), 7.55 (ddd, J = 8.2, 6.8, 1.4 Hz, 1H, H-7), 7.68 (dd, J = 7.9, 1.4 Hz, 1H, H-5), 7.68 (ddd, J = 7.9, 1.4, 0.5 Hz, 1H, H-6), 8.50 (dd, J = 8.2, 0.5 Hz, 1H, H-8), 9.99 (br s, 1H, 3-OH), 14.40 (s, 1H, 1-OH), HR-TOF-MS (ESI, +ve) m/z: 245.0266 [M+H]+ (calcd 245.0266 for C13H8O3S +H).
Preparation of 1,3-diallyloxyxanthone Analogues
To a stirred suspension of 1,3-dihydroxyxanthone analogues (2 mmol), allyl bromide (0.5 mL, 994 mg, 4.9 mmol) and K2CO3 (750 mg 5.4 mmol) in dry DMF for 24 hr. The reaction was monitored by TLC. After the reaction was completed, the reaction was diluted in EtOAc (25 mL) and washed with water (3 x 15 mL). The organic layers were collected and dried over Na2SO4. Evaporation afforded a yellow residue. Next the residue was purified by flash column chromatography (10% acetone-hexane) to give desired products in 64-75 % yield. 1,3-diallyloxyxanthone (3a), pale yellow needle, 74.6% yield, m.p.: 98-100°C, Rf: 0.35 (30% acetone-hexane), 1H NMR [CDCl3] δ: 4.59 (d, J = 5.26 Hz, 2H, H-1¢¢), 4.65 (d, J = 5.26 Hz, 2H, H-1′), 5.35 (d, J = 5.84 Hz, 1H, H-3′b), 5.29-5.34 (m, 1H, H-3”b), 5.68 (d, J = 17.28 Hz, 1H, H-3¢a), 5.43 (d, J = 17.24 Hz, 1H, H-3¢”a), 5.95-6.19 (m, 2H, H-2′), 5.95-6.19 (m, 2H, H-2”), 6.34 (s, 1H, H-2), 6.46 (s, 1H, H-4), 7.19-7.36 (m, 2H, H-5), 7.19-7.36 (m, 2H, H-7), 7.59 (t, J = 7.76 Hz, 1H, H-6), 8.25 (d, J = 7.92 Hz, 1H, H-8), HR-TOF-MS (ESI, +ve) m/z : 309.1197 [M+H]+ (calcd 309.1121 for C19H16O4+H).
1,3-diallyloxythioxanthone (5a), yellow solid, 64.3 % yield, m.p.: 60-61°C, Rf: 0.55 (30% acetone-hexane), 1H NMR [CDCl3] δ: 4.59 (d, J = 7.9 Hz, 1H, H-1”), 4.68 (d, J = 4.8 Hz, 1H, H-1′), 5.35 (dd, J = 10.5, 1.5 Hz, 1H, 3′b), 5.35 (dd, J = 10.5, 1.3 Hz, 1H, H-3”b), 5.64 (dd, J = 17.2, 1.5 Hz, 1H, 3′a), 5.64 (dd, J = 17.2, 1.3 Hz, 1H, H-3”a), 6.09 (m, 1H, 2′), 6.09 (m, 1H, H-2”), 6.46 (d, J = 2.3 Hz, 1H, H-2), 6.56 (d, J = 2.3 Hz, 1H, H-4), 7.42 (m, 1H, H-5), 7.46 (t, 1H, H-6), 7.42 (m, 1H, H-7), 8.46 (d, J = 7.9 Hz, 1H, H-8), HR-TOF-MS (ESI, +ve) m/z : 325.0913 [M+H]+ (calcd 325.0892 for C19H16O3S +H).
Claisen Rearrangement of 1,3-diallyloxyxanthone Analogues 3a and 5a
1,3-diallyloxyxanthone analogues (5 mmol) were dissolved in N,N-Dimethylformamide (DMF, 8 mL) and refluxed for 12 hr in an sand-bath. The reaction mixture was cooled at room temperature and quenched with dil. HCl (10%, 15 mL). The crude product was extracted with EtOAc (3 x 15 mL). The organic solution was dried, evaporated and chromatographed on silica gel by eluting with hexane-acetone (15% acetone-hexane) to give rearrangement products 3b, 5b, 3c and 5c in about 45-60% yield.
2,4-diallyl-1,3-dihydroxyxanthone (3b), pale yellow needle, m.p.: 136-138°C, Rf: 0.50 (30% acetone-hexane), 1H NMR [CDCl3] δ: 3.53 (d, J = 6.14 Hz, 2H, H-1”), 3.62 (d, J = 6.10 Hz, 2H, H-1′), 4.97-5.40 (m, 4H, H-3′a), 4.97-5.40 (m, 4H, H-3”a), 4.97-5.40 (m, 4H, H-3”b), 4.97-5.40 (m, 4H, H-3′b), 5.89-6.08 (m, 2H, H-2′), 5.89-6.08 (m, 2H, H-2”), 6.10 (s, 1H, 3-OH), 7.35 (t, J = 7.55 Hz, 1H, H-7), 7.44 (d, J = 8.39 Hz, 1H, H-5), 7.70 (ddd, J = 8.39, 1.2 Hz, 1H, H-6), 8.24 (dd, J = 7.9, 1.2 Hz 1H, H-8), 13.25 (s, 1H, 1-OH), HR-TOF-MS (ESI, +ve) m/z : 309.1124 [M+H]+ (calcd 309.1121 for C19H16O4 +H).
2,4-diallyl-1,3-dihydroxythioxanthone (5b), pale yellow needle, m.p.: 98-100°C, Rf: 0.45 (30% acetone-hexane), 1H NMR [CDCl3] δ: 3.57 (d, J = 5.9 Hz, 1H, H-1′), 4.62 (d, J =4. 7 Hz, 1H, H-1”), 5.08 (d, J = 12.0 Hz, 1H, H-3”b), 5.16 (d, J =26.1 Hz, 1H, H-3¢¢a), 5.17 (d, J = 17.4 Hz, 1H, H-3’b), 5.22 (d, J = 25.7 Hz, 1H, H-3′a), 6.08 (s, 1H, 3-OH), 6.95 (m, 1H, H-2′), 6.95 (m, 1H, H-2”), 7.43 (t, J = 8.0 Hz, 1H, H-7), 7.55 (m, 1H, H-5), 7.55 (m, 1H, H-6), 8.53 (d, J = 8.0 Hz, 1H, H-8), 15.13 (s, 1H, 1-OH), HR-TOF-MS (ESI, +ve) m/z : 325.0896 [M+H]+ (calcd. 325.0892 for C19H16O3S +H).
2-allyl-3-allyloxy-1-hydroxyxanthone (3c), yellow solid, m.p.: 118-200°C, Rf: 0.62 (30% acetone-hexane), 1H NMR [CDCl3] δ: 3.45 (d, J = 6.22 Hz, 2H, H-1′) 4.64 (d, J = 4.90 Hz, 2H, H-1”), 4.96 (dd, J = 10.0, 1.8 Hz, 1H, H-3′b), 4.96 (d, J = 10.0, 1.6 Hz, 1H, H-3”b), 5.04 (dd, J = 17.1, 1.8 Hz, 1H, H-3′a), 5.44 (dd, J = 17.1, 1.6 Hz, 1H, H-3”a), 5.88-6.13 (m, 2H, H-2′), 5.88-6.13 (m, 2H, H-2”), 6.42 (s, 1H, H-4), 7.35 (t, J = 7.5 Hz, 1H, H-7), 7.41 (d, J = 7.5 Hz, 1H, H-5), 7.68 (ddd, J =7.5,1.2 Hz, 1H, H-6), 8.24 (dd, J = 7.9, 1.2 Hz, 1H, H-8), 12.95 (s, 1H, 1-OH), HR-TOF-MS (ESI, +ve) m/z: 309.1129 [M+H]+ (calcd. 309.1121 for C19H16O4+H).
2-allyl-3-allyloxy-1-hydroxythioxanthone (5c), yellow solid, m.p.: 109-100°C Rf: 0.50 (30% acetone-hexane), 1H NMR [CDCl3] δ: 3.48 (d, J = 6.1 Hz, 1H, H-1′), 4.62 (d, J =4. 7 Hz, 1H, H-1”), 4.96 (d, J = 9.5 Hz, 1H, H-3′b), 5.05 (d, J = 17.1 Hz, 1H, H-3′a), 5.31 (d, J = 10.5 Hz, 1H, H-3”b), 5.44 (d, J = 17.2 Hz, 1H, H-3”a), 6.00 (m, 1H, H-2′), 6.00 (m, 1H, H-2”), 6.51 (s, 1H, H-4), 7.45 (m, 1H, H-5), 7.45 (m, 1H, H-7), 7.56 (t, J =7.3 Hz, 1H, H-6), 8.44 (d, J = 7.7 Hz, 1H, H-8), 14.15 (s, 1H, 1-OH), HR-TOF-MS (ESI, +ve) m/z: 325.0894 [M+H]+ (calcd. 325.0892 for C19H16O3S+H).
In Vitro Xanthine Oxidase Inhibitory Activity
The inhibitory activity of each compound was determined using a slight modification of the reference methods12. The assay mixture consisted of 10 mL of the tested compound (18.75, 37.5, 75.0, 150.0, 300.0 and 600.0 μg/mL), 180.0 mL of phosphate buffer (pH 7.5) and 10 mL of 0.06 mM xanthine solution (in phosphate buffer, pH 7.5). The assay reaction was started by the addition of 10 mL of xanthine oxidase enzyme solution (0.4 units/mL in phosphate buffer, pH 7.5), which was prepared immediately before use. The assay mixture was incubated at 37ºC for 30 min. The reaction was stopped by adding 10 mL of 0.5 M HCl. For blank assays, the protocol is similar but adding 0.5 M HCl before enzyme. The absorbance at 295 nm was recorded using a microplate reader (BioTek Synergy HTX). The test was performed in triplicate and allopurinol (5 – 100 μg/mL) was used as the standard.
Computational Studies
The three-dimensional structure of xanthine oxidase from bovine milk was obtained from Protein Databank (www.pdb.org, PDB code 1FIQ) from 2.50 Å resolved crystal structure13. Before molecular docking calculations, the crystallographic water molecules in the structure were removed, and the galantamine inhibitor was separated. The ligands were docked into the salicylic acid binding site of XO using AutoDock 4.214. AutoDock 4.2 in Linux version was used because its algorithm allows full flexibility of small ligands. The best conformers were searched by the Lamarckian genetic algorithm (LGA). During the docking process, a maximum of 100 conformers was considered. The population size was set to 150. A maximum number of energy evaluation was set to 2500000. A grid box size of 100 Å x 100 Å x 100 Å with a spacing of 0.375 Å between the grid points and centered at 33.836 (x), 8.227 (y) and 30.991 (z) was used covering the active site of the 1FIQ. The lowest energy cluster returned by AutoDock for the compound was used for further analysis. All other parameters were maintained at their default settings. Before docking, the ligands were optimized at the HF/3-21G level of calculation using Gaussian 03 packages15. All molecular graphics material was prepared using the Discovery Studio 3.1 Client (ver. 3.1.1.11157, Accelrys Software Inc., San Diego, CA, USA). Docking method validation was done by redocking natural ligand receptor on the active site. The docking validation method was evaluated based on the value root-mean-square deviation (RMSD) and declared valid if the value of RMSD smaller than 3.5 Å16. In this study, the RMSD was 0.88 indicating this protocol and calculation parameter settings meet the criteria of validity docking methods.
Results and Discussion
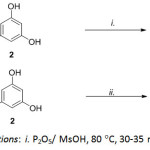 |
Scheme 1: Reaction conditions: i. P2O5/ MsOH, 80 °C, 30-35 min; ii. MsOH, Al2O3, 110°C. Click here to View scheme |
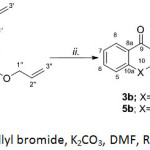 |
Scheme 2: Reaction conditions: i. Allyl bromide, K2CO3, DMF, RT, 24 hr.; ii. DMF, reflux, 24 hr. Click here to View scheme |
Chemistry
1,3-Dihydroxyxanthone (3), was prepared in 71% yield by heating a mixture of salicylic acid (1) and phloroglucinol (2) using a freshly prepared mixture of methane sulfonic acid and P2O5 under heating at 80oC (Scheme 1A). In a likewise manner, 1,3-dihydroxythioxanthone (5) were obtained from treating thiosalicylic acid (4) and phloroglucinol (2) in alumina acidic type-methanesulfonic acid which served as a catalyst and also as a coupling agent to give the thioxanthone building block (Scheme 1B). The synthesis of di-O-allyl xanthone derivatives 3a and 5a were carried out by the interaction of allyl bromide with corresponding hydroxyxanthone derivatives in the presence of K2CO3. After the completion of etherification, compound 3a and 5a were subjected to Claisen rearrangement under thermal conditions smoothly provided 3b-3c and 5b-5c. The structures of synthesis compounds were confirmed mainly using NMR (1D- and 2D-NMR) and HR-MS techniques. Spectroscopic data of 3-5 were in agreement with the structures and were consistent with the literature values.
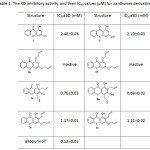 |
Table 1: The XO inhibitory activity and their IC50 values (µM) for xanthones derivatives Click here to View table |
Biological Activity
The inhibition of xanthine oxidase results in a decreased production of uric acid was measured spectrophotometrically at 295 nm. All synthesized compounds were evaluated for XO inhibitory activity and their IC50 values are shown in Table 1. Among the compounds tested hydroxylated xanthone series, 2,4-diallyl-1,3-dihydroxythioxanthone (5b, IC50 = 0.69±0.02 mM) was most active which was comparable to that of 2,4-diallyl-1,3-dihydroxyxanthone (3b, IC50 = 0.76±0.03 mM). It was less active than the standard gout drug allopurinol (6, IC50 = 0.12±0.01 mM). A drop-in activity was noted for 3c and 5c with their IC50 1.17±0.01 and 1.11±0.02 mM, respectively. The parent compounds, 3 and 5, displayed less active than their derivatives with IC50 2.48±0.03 mM and 2.19±0.03 mM, respectively. However, the loss of inhibitory potency was observed in di-O-allyl derivatives 3a and 5a. It should be noted that the hydroxyl groups at C-1 and C-3 seemed crucial as was evident from the activity of 3b and 5b. Based on potent XO inhibitory activity of 5b, the compound 5b was selected for further molecular modeling study
Molecular Modeling
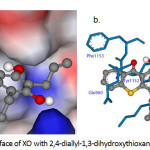 |
Figure 1a: Molecular surface of XO with 2,4-diallyl-1,3-dihydroxythioxanthone (5b). Click here to View figure |
Selected Molecular Interaction of Compound 5b in the Binding Sites of XO
Molecular docking study was performed to comprehend the binding mode of synthesized inhibitor and XO. To further investigate the interaction mode of 2,4-diallyl-1,3-dihydroxythioxanthone (5b) to XO (1FIQ). The conformation with the least binding energy, having maximum cluster size at the active site of the respective protein was considered for interaction studies. As shown in Table 2 and Fig.1, 2,4-diallyl-1,3-dihydroxythioxanthone (5b) showed binding energy -9.50 kcal/mol. The predicted binding mode of the highest active compound against the XO model displayed hydrogen bond interactions occurred with amino acids residues. Six hydrogen bond interactions of the highest active compound with Glu960, Phe1153, Pro1248, Asn1249 and Lys1250 residues of the 1FIQ were observed. The first hydrogen bond was observed between the 1-OH of 5b and Glu960 residue (4.061 Å). The second and the third hydrogen bond were found between the C-1 hydroxyl group and the C-9 carbonyl group of 5b and Phe1153 distance 3.658 and 3.184 Å, respectively. The forth hydrogen bond were detected between the C-3 hydroxyl group of 5b and Pro1248 (3.066 Å). Also, two H-bond interactions between C-3 hydroxyl group and Asn1249 (3.066 Å) and Lys1250 (3.092 Å) were detected. The allyl moieties of 5b interacted with Lys1250 (3.150 Å) and Lys1251 (3.214 Å) residues via hydrophobic interactions. Compound 5b formed O-S interaction with the residues Glu960 (3.404 Å). Furthermore, two O-C interactions were found with the residues Asp1181 and Cys1247 distance 3.610 and 3.020 Å, respectively. All these interactions helped 5b to hold in the binding site of the XO.
Table 2: The interactions data for compound 5b docked into binding site of XO (1FIQ)
|
Ligand |
Residue |
Type of interaction |
Distance (Å) |
Ligand interacting moiety |
|
5b (binding energy -9.50 kcal/mol) |
Glu960 |
Hydrogen bond |
4.061 |
1-OH |
|
O-S interaction |
3.404 |
S |
||
|
Tyr1152 |
C-C interaction hydrophobic |
3.256 |
C-8 |
|
|
Phe1153 |
Hydrogen bond |
3.658 |
1-OH |
|
|
Hydrogen bond |
3.184 |
C=O |
||
|
C-C interaction hydrophobic |
3.560 |
C-8 |
||
|
Asp1181 |
O-C interaction |
3.610 |
3’b |
|
|
Cys1247 |
O-C interaction |
3.020 |
3’b |
|
|
Pro1248 |
Hydrogen bond |
3.066 |
3-OH |
|
|
Asn1249 |
Hydrogen bond |
3.066 |
3-OH |
|
|
Lys1250 |
Hydrogen bond |
3.092 |
3-OH |
|
|
C-C interaction hydrophobic |
3.150 |
C-2”b |
||
|
Lys1251 |
C-C interaction hydrophobic |
3.214 |
C-2”’b |
Conclusions
In this study, a series of hydroxyxanthones and hydroxythioxanthones have been synthesized and evaluated for the inhibition of XO activity. Among the tested, 2,4-diallyl-1,3-dihydroxythioxanthone (5b) had a highly inhibitory effect on XO with IC50 = 0.69±0.02 mM as compared to standard allopurinol (IC50 = 0.12±0.01 mM). Molecular docking study was fulfilled to understand the molecular interaction of compounds with the active site of XO. This study has identified a new series of lead compounds which can be used for the development of new XO inhibitors for the prevention and treatment of gout.
Acknowledgments
The authors are grateful to Faculty of Science and Technology, Phranakhon Rajabhat University for providing necessary research facilities and we would also like to thank Buker daltonic LLC for providing ESI-HR-TOF-MS.
References
- Cejková, J.;Ardan, T.; Filipec, M.; Midelfart A. Histol. Histopathol. 2002,17(3),755-760.
- Pacher, P.; Nivorozhkin, A.; Szabó, C.; Pharmacol. Rev. 2006, 58(1), 87.
CrossRef - Lonjou, C.; Borot, N.; Sekula, P.; Ledger, N.; Thomas, L.; Halevy, S.; Naldi, L.; Bouwes-Bavinck, J.-N.; Sidoroff, A.; de Toma, C.; Schumacher, M.; Roujeau, J.-C.; Hovnanian, A.; Mockenhaupt, M. Pharmacogenet. Genomics. 2008, 18(2), 99-107.
CrossRef - Cicerale, S.; Lucas, L.; Keast, R.; Int. J. Mol. Sci. 2010, 11(2), 458-479.
CrossRef - Peres, V.; Nagem, T.J.; de Oliveira, FF. Phytochemistry. 2000, 55(7), 683-710.
CrossRef - Hu, L.; Hu, H.; Wu, W.; Chai, X.; Luo, J.; Wu, Q. Bioorg. Med. Chem. Lett. 2011, 21(13), 4013-4015.
CrossRef - Rosi, D.; Peruzzotti, G.; Dennis, E.W.,; Berberian, D. A.; Freele, H.; Tullar, B. F. Archer, S. J Med Chem. 1967, 10(5), 867-876.
CrossRef - Luerang, W.; Khammee, T.; Kumpum, W.; Suksamrarn, S.; Chatsudthipong, V.; Muanprasat, C.; Life Sci. 2012, 90(25-26), 988-994.
CrossRef - Coelho, P.J.; Carvalho, L.M.; Silva, J.C.; Oliveira-Campos A.M.F.; Samat, A.; Guglielmetti, R. Helv. Chim. Acta. 2001, 84(1), 117-123.
CrossRef - Pillai, R.K.M.; Naiksatam, P.; Johnson, F.; Rajagopalan, R.; Watts, P. C.; Cricchio, R.; Borras, S. J. Org. Chem. 1986, 51(5), 717-723.
CrossRef - Sharghi, H.; Beni, A.R.S. Synthesis. 2004, 2004, 2900-2904.
- Kong, L.D.; Zhang, Y.; Pan, X.; Tan, R.X.; Cheng, C.H.K., Cell. Mol. Life. Sci. 2000, 57(3), 500-505.
CrossRef - Enroth, C.; Eger, B.T.; Okamoto, K.; Nishino, T.; Pai, E.F. Proc. Natl. Acad. Sci. U S A 2000, 97(20),10723-10728.
CrossRef - Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M. F.; Belew, R. K.; Goodsell, D. S. Olson, A. J. J. Comput. Chem. 2009, 30(16), 2785-2791.
CrossRef - Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G. E.; Robb, M. A.; Cheeseman, J. R.; Montgomery, J. A.; Vreven, T.; Kudin, K. N.; Burant, J. C.; Millam, J. M.; Iyengar, S. S.; Tomasi, J.; Barone, V.; Mennucci, B.; Cossi, M.; Scalmani, G.; Rega, N.; Petersson, G. A.; Nakatsuji, H.; Hada, M.; Ehara, M.; Toyota, K.; Fukuda, R.; Hasegawa, J.; Ishida, M.; Nakajima, T.; Honda, Y.; Kitao, O.; Nakai, H.; Klene, M.; Li, X.; Knox, J. E.; Hratchian, H. P.; Cross, J. B.; Bakken, V.; Adamo, C.; Jaramillo, J.; Gomperts, R.; Stratmann, R. E.; Yazyev, O.; Austin, A. J.; Cammi, R.; Pomelli, C.; Ochterski, J. W.; Ayala, P. Y.; Morokuma, K.; Voth, G. A.; Salvador, P.; Dannenberg, J. J.; Zakrzewski, V. G.; Dapprich, S.; Daniels, A. D.; Strain, M. C.; Farkas, O.; Malick, D. K.; Rabuck, A. D.; Raghavachari, K.; Foresman, J. B.; Ortiz, J. V.; Cui, Q.; Baboul, A. G.; Clifford, S.; Cioslowski, J.; Stefanov, B. B.; Liu, G.; Liashenko, A.; Piskorz, P.; Komaromi, I.; Martin, R. L.; Fox, D. J.; Keith, T.; Laham, Al; Peng, C. Y.; Nanayakkara, A.; Challacombe, M.; Gill, P. M. W.; Johnson, B.; Chen, W.; Wong, M. W.; Gonzalez, C.; Pople, J. A. Gaussian 03, Revision C.02, In, 2003.
- Kontoyianni, M.; McClellan, L.M.; Sokol, G.S. J Med Chem. 2004, 47(3) 558-565.
CrossRef

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.

This work is licensed under a Creative Commons Attribution 4.0 International License.









