Nanocrystalline Mixed Ligand Complexes of Cu (II), Ni (II), Co (II) with N, O Donor Ligands: Synthesis, Characterization, and Antimicrobial Activity
Kolhe Nitin H, Jadhav Shridhar S, Shaikh Sajid H, Takate Sushma J and Aware Dinkar J.
1Department of Chemistry and Research Centre , New Arts Commerce and Science College Ahmednagar; affiliated to Savitribai Phule Pune University Lal-taki Rd -414401, Ahmednagar (M.S.) India.
2Department of Microbiology, New Arts Commerce and Science College, Ahmednagar.
3Department of Chemistry, Arts, Commerce and Science College Sonai, Tal-Newasa,Dist.-Ahmednagar -414603,affiliated to.S. P.Pune University,India.
Corresponding Author E–mail: ssjadhav1957@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/320620
In present investigation nanocrystalline mixed ligand complexes were synthesized using 8-hydroxyquinoline, salicylaldoxime with metals like Cu (II), Ni (II) and Co (II). The metal: ligand ratio was found to be 1:1:1. These complexes were characterized using electronic spectra, FTIR spectra, elemental analysis, magnetic susceptibility, thermogravimetric analysis, conductivity measurement, powder X-ray diffraction and Scanning Electron Microscopy with electron dispersive spectroscopic methods. The electronic spectra of complexes suggest that they have square planer geometries. In FTIR analysis characteristic bands of ν (M-N) and ν (M-O).The Co (II) and Cu (II) complexes are paramagnetic in nature and these had square planer geometry. While Ni (II) complexes are diamagnetic nature and having square planer geometry. The thermal analysis of complexes was studied in an attempt to assign intermediate compounds. Low molar conductance values indicate non – electrolytic nature of the complexes. The Powder X-ray diffraction study shows formation of nanocrystalline phase as well as the grain size of complexes is less than 10 nm. The EDS study is shows good agreement for formation of mixed ligand metal complexes . Complex: [C16H12CuN2O2], [C14H12N2NiO4 ], [C16H12NiN2O2] and [C16H12CoN2O2] had antimicrobial activity against four bacteria tested. Bacteria were resistant to other five complexes.
KEYWORDS:Nanocrystalline complexes; N; O donor; SEM-EDS; Disc Diffusion
Download this article as:| Copy the following to cite this article: Nitin H. K, Shridhar S. J, Sajid H. S, Sushma J. T, Dinkar J. A. Nanocrystalline Mixed Ligand Complexes of Cu (II), Ni (II), Co (II) with N, O Donor Ligands: Synthesis, Characterization, and Antimicrobial Activity. Orient J Chem 2016;32(6). |
| Copy the following to cite this URL: Nitin H. K, Shridhar S. J, Sajid H. S, Sushma J. T, Dinkar J. A. Nanocrystalline Mixed Ligand Complexes of Cu (II), Ni (II), Co (II) with N, O Donor Ligands: Synthesis, Characterization, and Antimicrobial Activity. Orient J Chem 2016;32(6).Available from: http://www.orientjchem.org/?p=25851 |
Introduction
A lot of attention is being given on the study of simple transition metal complexes and their biological activity1. The N, O donor ligands have been used as chelating agents in the field of coordination chemistry and medicine. The importance of N, O donor ligands and their metal complexes in different biological activities is very well known2. These biological activities include anti-inflammatory, antimicrobial, anticancer and antidermal activity. They are also studied as enzyme inhibitors. Cis-platin is one of the best examples of mixed ligand complex.3,4.The Nano crystalline quantum dot mixed ligands complexes play an important role in inorganic, organometallic chemistry and medicinal chemistry. A lot of research has been carried out on synthesis and characterization of mixed ligand transition metal complexes.5–6. But till date nanocrystalline complexes of transition metal ion have been given very little attention. Therefore study of nanocrystalline mixed ligand transition metal complexes has a great scope in the field of research in inorganic chemistry. Due to the change in their physical properties, such as surface area, decomposition temperature, magnetic properties, these nanocrystalline metal complexes can be used as catalyst in organic synthesis .Considering all above facts the aim of current research is synthesis and characterization of nanocrystalline mixed ligand complexes of Co (II), Ni(II) and Cu (II) with N,O donors and study of their antimicrobial activity.
Experimental
Chemicals
AR grade metal salts of NiCl2.6H2O, CuCl2.2H2O andCoCl2.6H2O were purchased from Loba chemicals. The salicylaldoxime and 8-hydroxyquinoline, ethanol, potassium hydroxide were purchased from Merck chemicals.
General procedure for synthesis of complexes Ni(II), Cu(II) and Co (II) ions with salicylaldoxime ligands
1 mmole of salicylaldoxime was dissolved in alcoholic KOH .This solution was added drop wise to the solution of 1mmole metal salts in alcohol. The reaction mixture was refluxed for appropriate time. The solid product was obtained after completion of the reaction. The Co (II) with ligand reaction was kept in inert atmosphere and solution was bubbled with nitrogen gas for 10 minutes. All the complexes of salicylaldoxime with Ni(II), Cu(II) ions and Co (II) were synthesized by reported methods.7
General Procedure for Synthesis of complexes of N, O (8-Hydroxyquinoline) donors with Ni (II), Cu (II) and Co(II) ions
The ligand 8-Hydroxyquinoline with Ni (II), Cu (II) and Co (II) ions was synthesized by modifying literature methods8.1mmole of 8-hydroxyquinoline was dissolved in alcoholic KOH. This solution was added drop wise to the solution of 1mmole metal salt in alcohol. The reaction mixture was refluxed for appropriate time. The solid product was obtained after completion of the reaction.
Synthesis of mixed ligand complexes of 8-hydroxyquinoline and salicylaldoxime with Ni (II), Cu (II) and Co (II) metal ions
1mmole of metal salts was dissolved in alcohol. To this 1mmole of 8-hydroxyquinoline and 1mmole of salicylaldoxime in alcoholic KOH were added respectively with constant stirring. The mole ratio of M :L1:L2 was 1:1:1 and reaction mixture was refluxed for 6 hrs. In synthesis of Co (II) complex the reaction was carried out in inert atmosphere to prevent the oxidation of Co(II). The reaction mixture was evaporated to about half of the original volume which on cooling afforded the solid product. The product was filtered, washed with cold dry ethanol and dried in vacuum.
Antimicrobial activity of Complexes and Ligands
The antimicrobial activities of ligand as well as complexes were tested by agar disc diffusion method using Mueller Hinton Agar (MHA).9,10 Antimicrobial activity of each complex was tested for Bacillus subtilis NCIM 2063 and Escherichia coli NCIM 2341. The MHA was prepared and poured in the plates after sterilization. The plates were allowed to solidify for 15 minutes. Sterile 5mm Whatmann filter paper disc was loaded with test compounds (100μg/disc) and placed over inoculated plates. The dimethylsulfoxide (DMSO) was kept as negative control.11 The plates were then incubated at 37ºC for 24h. After incubation the plates were observing for zones of inhibition. The diameter of zones of inhibition was recorded for the positive plates.
Physical Measurements and analysis
The FTIR spectra of complexes were obtained on Shimadzu IRAffinity-1 Fourier transform Infrared spectrophotometer on samples pressed on KBr pellets.UV-Vis absorption measurements were carried out on a Shimadzu UV1800at 26 ºC in 10-3 in DMF and DMSO. The Elemental analysis (C, H, and N) was done by using PerkinElmer analyzer. Magnetic properties were determined from Gouy balance method. The thermogravimetric analysis was carried out on Shimadzu TGA50H thermoanalyzer. The synthesized nanocrystalline powders were characterized by various sophisticated techniques such as powder X-ray diffraction (XRD) patterns recorded on a model Philips X-ray diffract meter with diffraction angle 2θ in between 20° to 80° using Cu-Kα radiation of wavelength 1.54058 Å. The grain size was calculated from grain size programmer software by using the XRD data such as FWHM, wavelength of Cu-Kα radiation. The Surface morphology and elemental analysis of the samples were carried out using scanning electron microscopy with electron dispersion spectroscopy (SEM-EDS) characterization conducted using a JEOL-JEM -6360A Analytical scanning Electron Microscope(Oxford).
Results and Discussion
The elemental analysis data, decomposition temperature, colour, mol. wt., percentage yield and molar conductance were given in Table 1.
Table 1: Physical Characteristics and Analytical data for ligands and their metal complexes
|
Sr.No. |
Complexes |
Color |
Mol. Wt. |
M.P 0C |
Yield % |
Elemental analysis |
Molar Conductance Ω-1cm2 mol-1 |
|||
| C | H | O | N
|
|||||||
| L1 | [C9H7NO] | White | 145.15 | 74 | — | — | — | — | — |
— |
| L2 | [C7H7NO2] | White | 137.13 | 58 | — | — | — | — | — |
— |
| 1 | [C14H12 CuN2O4 ] | Yellowish Green | 335.80 | 246d* | 74.55% | 50.07(49.80) | 3.00(2.97) | 19.76(19.68) | 8.34(8.33) |
06.30 |
| 2 | [C18H12CuN2O2 ] | Faint Green | 351.84 | 256d* | 71.21% | 61.45(61.39) | 3.44(3.30) | 9.09(9.10) | 7.96(7.89) |
09.60 |
| 3 | [C16H12CuN2O3] | Green | 343.83 | >300d* | 68.00% | 55.89(55.81) | 3.52(3.27) | 13.16(12.98) | 8.15(8.11) |
03.75 |
| 4 | [C14H12N2NiO4 ] | Greenish Yellow | 330.94 | 135d* | 67.34% | 50.81(50.54) | 3.65(3.64) | 19.34(19.32) | 8.46(8.44) |
07.35 |
| 5 | [C18H12NiN2O2 ] | Green | 346.99 | 263d* | 53.00% | 62.30(62.28) | 3.49(3.48) | 9.22(9.22) | 8.07(8.07) |
07.70 |
| 6 | [C16H12NiN2O3] | Dark olive Green | 338.97 | 230d* | 65.63% | 56.69(56.63) | 3.57(3.49) | 14.16(13.69) | 8.16(8.07) |
09.26 |
| 7 | [C14H12CoN2O4 ] | Radish Brown | 331.18 | >305d* | 58.06% | 56.20(56.18) | 4.04(4.04) | 10.70(10.68) | 9.36(9.30) |
01.90 |
| 8 | [C18H12CoN2O2 ] | Brown | 347.23 | >300d* | 59.93% | 62.26(62.24) | 3.48(3.47) | 9.22(9.19) | 8.07(8.05) |
01.65 |
| 9 | [C16H12CoN2O3] | Chocolate Brown | 339.21 | 290d* | 57.74% | 56.65(56.62) | 3.57(3.55) | 14.15(14.10) | 8.26(8.11) |
03.75 |
d* = Decomposition temperature
Infrared Spectroscopy
The FTIR spectra provide valuable information on the nature of the functional group attached to the metal atom. The characteristic IR frequencies of ligand 8-hydroxyquinoline (L1) [C9H7NO], salicylaldoxime (L2) [C7H7NO2 ] and their complexes were described in Table 2. The FTIR Spectrum of C9H7NO (L1) shows the frequency 3300-3340 cm-1 is attributed to the phenolic OH group. The frequency 1412cm-1 attributed to C=N in the ring12 and the N-H frequency 3300-3600 cm-1. The infrared spectrum of the [C7H7NO2 ] ligand showed strong and broad band’s due to the hydrogen bonded phenolic OH at O-position in the region 3000–2800 cm–1. It also exhibits two separate OH bands of the oxime OH at 3237cm-1 and 3137 cm–1 and phenolic OH at 3400 cm–1. The dwarf bands observed in the 1646–1620 cm–1 frequency ranges of the complexes were assigned to the ν(C=N).13 The shift of the ν(C=N) vibration in all the complexes to a lower frequency suggests that the nitrogen atom of the ring contributes to the complexation. The lower ν(C=N) frequency also indicates stronger M–N co-ordination bonding. In the IR spectra of the complexes, a band was observed between 403cm-1 and 476 cm−1, which is attributed to ν(M–N) stretching vibrations.14 Another band appeared between 493cm-1 and 619 cm−1, which are assigned to the interaction of the phenolic oxygen to the metal atom.
In all the Cu(II) metal ligand complexes, a band appear in the range of 403cm−1to 476cm−1is attributed to ν(Cu-N).15–16 In case of [C18H12CuN2O2] shows 403 cm−1, while [C14H12 CuN2O4] shows 476 cm−1. The complex [C16H12CuN2O3] shows νCu-O frequency at 466 cm−1is in between the [C18H12CuN2O2] and [C14H12CuN2O4] indicate the formation of the mixed ligand complex. The ν(Cu-N) for the complexes [C18H12CuN2O2], [C14H12CuN2O4 ] and [C16H12CuN2O3] is 596cm−1, 525 cm−1and 518cm−1, respectively. This observation is similar to that of ν(Cu-O) for all the copper complexes. Similarly the ν(C=C) and ν(C-O) for these complexes were at 1446cm−1, 1469 cm−1, 1475cm−1 and 1545 cm−1, 1139cm−1,1597cm−1respectively. The frequency of ligand [C9H7NO] shows 1500cm−1 and 1215cm−1is attributed to νC=C and νC-O- respectively. The decreased frequency in Cu (II)-Ligands complex by 54 cm−1to 25 cm−1 and 76 cm−1units indicates formation of complex.
In case of [C14H12N2NiO4], [C18H12NiN2O2] and [C16H12NiN2O3] complexes, the bands at 426cm−1, 450 cm−1,465cm−1 and 508cm−1,505cm−1 and 504cm−1were attributed to ν(Ni-N) and ν(Ni-O) bond formation between ligands and metal ion. The bands at 3059 cm−1, 3062 cm−1were attributed to aromatic –C=C- and 2955 cm−1 were attributed to ν(-C-H), Ni(II)-L complexes. The ν(C-O-) and ν(C=N) frequencies observed to be decreased in complexes by 56 cm−1 and 90 cm−1 respectively as compared with ligands. Mainly the frequency of mixed ligands complexes shows the intermediate frequency between the metal complexes.
In case of all the three complexes of Co(II), ν(Co-O) ,ν (Co-N),ν(C=N), (C-O-) bands were observed at 410 cm−1, 462 cm−1, 450 cm−1, 493 cm−1,505 cm−1, 619cm−1, 1545cm−1, 1542cm−1,1639cm−1, 1469 cm−1,1423 cm−1 ,1420 cm−1 respectively.
Table 2: The characteristic infrared-absorption band of ligands and their complexes
| Sr.No. | Complexes | IR-Absorption band in cm-1 | |||||||
| ᶹO-H | ᶹC-H | ᶹC=O | ᶹC=N | ᶹC=C | ᶹC-O | ᶹM-N | ᶹM-O | ||
| L1 | [C9H7NO] | 3213 | 3074 | —- | —- | 1500 | 1215 | —- | —- |
| L2 | [C7H7NO2] | 3209 | 2974 | —- | 1425 | —– | —- | —- | —– |
| 1 | [C14H12 CuN2O4 ] | —— | 3016 | —– | 1599 | 1446 | 1545 | 476 | 596 |
| 2 | [C18H12CuN2O2 ] | 3381 | 2708 | 1726 | 1498 | 1469 | 1139 | 403 | 525 |
| 3 | [C16H12CuN2O3] | —- | 3012 | —– | 1599 | 1475 | 1597 | 466 | 518 |
| 4 | [C14H12N2NiO4 ] | ——- | 3059 | 1643 | 1552 | 1444 | 1251 | 426 | 508 |
| 5 | [C18H12NiN2O2 ] | —— | 3062 | — | 1500 | 1487 | 1234 | 450 | 505 |
| 6 | [C16H12NiN2O3] | —— | 2955 | 1639 | 1520 | 1410 | 1200 | 465 | 504 |
| 7 | [C14H12CoN2O4 ] | ——- | 3066 | 1649 | 1545 | 1469 | 1251 | 410 | 493 |
| 8 | [C18H12CoN2O2 ] | ——- | 3059 | 1602 | 1542 | 1423 | 1238 | 462 | 505 |
| 9 | [C16H12CoN2O3] | —— | 2955 | 1600 | 1639 | 1420 | 1255 | 450 | 619 |
Electronic Spectra
The electronic spectra are shown in fig.1 for ligands and their metal complexes. The ligand band D for L1: [C9H7NO] and band B for L2 : [C7H7NO2] shows two absorption bands at 245nm, 275nm and 247nm, 265nm, respectively. However, it appears reasonable to assign the bands π → π* transitions due to π-bonded electron and n→ π* due to non-bonded pair of the electron on the oxygen atom respectively.17
The two bands were observed for heteroleptic complex of copper such as I: [C16H12CuN2O3] (Fig. 1).These bands were assigned with 2B2g→2B1g and 2Eg→2B1g transition at 575nm and 478nm respectively. These transitions indicate the geometry of complex is square planar [19]. It was also supported by magnetic susceptibility measurement. The other two homoleptic complex G: [C18H12CuN2O2 ], H:[C14H12 CuN2O4 ] shows absorption at 415nm and 500nm, at 345nm and 525nm respectively.
Complex F:[C16H12NiN2O2] exhibited three bands at 623nm, 428nm and 224nm .Which was attributed to the 3A2g→3T2g(F) (υ1); 3A2g→3T1g(F) (υ2) and 3A2g→3T1g(p) (υ3) transitions respectively .This spectral bands indicate the complex was square planar geometry. The complexes like C:[C18H12NiN2O2], E:[C14H12N2NiO4 ], shows absorption at 598nm, 445nm and 250nm, 595nm, 443nm and 275nm receptively.
The electronic spectral band for L: [C16H12CoN2O3] exhibited two characteristic absorption bands of complex at 475nm and 418nm .The complex J: [C18H12CoN2O2 ] and K:[C14H12CoN2O4 ] were showed two characteristic absorption bands at the 570 nm,435nm and 562nm, 440nm respectively corresponding to 4T1g(F)→4T2g(F)(υ1); 4T1g(F)→4T1g(P)(υ2) . These bands were the characteristics of square planar Co (II) complexes.
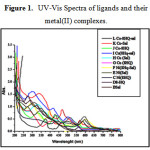 |
Figure 1: UV-Vis Spectra of ligands and their metal(II) complexes |
Magnetic Properties
The magnetic susceptibility measurements of the complex in the solid state was determined by Gouy balance using Hg [Co (SCN)4] as a calibrate. The magnetic moment of Cu (II)-complexes like [C14H12 CuN2O4 ], [C18H12CuN2O2 ], [C16H12CuN2O3] has been found to be μeff 1.83 BM., 1.81BM, 1.79BM indicative of one unpaired electron of Cu(II) ion and suggesting that the square-planar geometry of the complex.18,19.
All the Ni (II) complexes exhibited the diamagnetism phenomenon with square-planar geometry.20,21 The magnetic measurement of Co (II) complexes displayed magnetic moment value of 1.87B.M. 2.89BM, 2.34BM. This is the due to orbital contribution.22
Conductance Measurement
Conductance of the complexes is measured in DMF: H2O (40:60) solution at a concentration 10-4M. 23,24 The low molar conductance values of all metal complexes indicate that the complexes are non-electrolytes.25 The results of molar conductance are shown in Table1.
Thermogravimetric analysis of complexes
Thermogravimetric analysis Fig. 2a to2i of all the metals complexes was carried out to study the thermal stability and decomposition reaction as a function of time and temperature. In the present investigation, heating rate was suitably controlled at 5°C min-1 under non-isothermal way and the weight loss was measured from room temperature to the 1000°C26. The data is provided with in the Table 3.
Table 3: Thermoanalytical results TGA of Complexes
| Sr.No. | Complexes | Decomposition stages | Temp.rangeºC | Weight loss Found (Cal. )% | Decomposition assignment | Residue found(Cal.)% |
| 1 | [C14H12 CuN2O4 ] | Stage-1 | 198.59-207.59 | 59.46(60.15) | -C12H12O2, -N2 | CuO |
| Stage-2 | 428.208-607 | 16.74(19.43) | -C2H2 | |||
| 2 | [C18H12CuN2O2 ] | Stage-1 | 281.90-847.03 | 81.96(82.53) | – C18H12N2O2 | CuO |
| 3 | [C16H12CuN2O3] | Stage-1 | 213.47-951.69 | 88.38(88.50) | -NO, -C16H14NO | CuO |
| 4 | [C14H12N2NiO4 ] | Stage-1 | 135.12-176.05 | 14.67(15.62) | -N2H2 [O] | NiO |
| Stage-2 | 253.14-267.18 | 30.99(31.085) | -C7H3 | |||
| Stage-3 | 355.24-730.32 | 35.87(36.05) | -C7H2O | |||
| 5 | [C18H12NiN2O2 ] | Stage-1 | 311.27-989.38 | 81.30(83.07) | -C18H12N2O2 | NiO |
| 6 | [C16H12NiN2O3] | Stage-1 | 178.09-887.21 | 85.94 (86.78) | C16H12N2O | NiO |
| 7 | [C14H12CoN2O4 ] | Stage-1 | 199.93-202.94 | 59.167(60.23) | -C12H12O2,-N2 | CoO |
| Stage-2 | 383.33-433.38 | 17.71(20.16 ) | -C2H2 | |||
| 8 | [C18H12CoN2O2 ] | Stage-1 | 112.77-168.55 | 13.24 (14.15) | -NO,1/2O2 | CoO |
| Stage-2 | 397.56-592.93 | 56.01(57.03) | -C12H11N | |||
| 9 | [C16H12CoN2O3] | Stage-1 | 109.98-278.69 | 10.28(10.61) | -H2O, ½ O2 | CoO |
| Stage-2 | 387.04-401.46 | 46.052(47.76) | -C7H6NO |
The thermogram Fig. 2a for [C14H12 CuN2O4 ] shows that, the thermal decomposition of complex takes place in the temperature range 198.59°C -207.59 ºC and is accompanied by a weight loss of 59.46 % as against the calculated weight loss 60.15 % .These weight loss was due to the two phenolic groups and nitrogen gas evolved from the complex. The decomposition step is obtained in temperature range 428°C to 607 ºC. This step shows weight loss 16.74 %. This is in accordance with calculated weight loss 19.43 % due to loss of-C2H2 from the complex. The remaining formation of copper oxide as a residue.27
The thermogram fig. 2b for [C18H12CuN2O2] shows that [C18H12CuN2O2] shows that its decomposition takes place in temperature range 281.90°C – 847.03ºC and is equivalent to loss of two [C9H7NO] molecules by weight loss 81.96%, which is in accordance with calculated weight loss 82.53% .The end product, is the formation of CuO residue. The mixed ligand complex of Cu (II) –L1L2 shows very interesting result of TG. The decomposition temperature of this complex is in the range 213.43°C -951.69°C, which is in between Cu (L1) and Cu (L2) complexes. At the temperature 326.45°C loss of NO group and further decomposition reaction was continue to decompose the organic molecules .The total weight loss of this step is 88.38% which is in accordance to the calculated weight loss 88.50 % . This weight loss is equivalent to the of L1 and L2. The chocolate brown coloured CuO residue was obtained.
All the complexes of Ni(II) showed fig. 2d to 2f the final product NiO residue.28The complex [C14H12N2NiO4 ] thermogram shown in fig.2d decomposes in three steps. The weight loss observed was 14.67% in first step, 30.995% in second and 35.87% in the third step at 135.12°C -176.05°C, 253.14°C-267.18°C, 353.24°C -730.32 ºC, with loss N2H2O, C7H3, and C7H2O groups respectively. The TG of complex [C18H12NiN2O2] shows the decomposition of C18H12N2O2 organic compound at the 311.27°C -989.38 ºC .The % weight loss was found to be 81.30%. This was attributed to calculate weight loss was 81.07%. The mixed ligand complex of Ni (II) –L1L2 decomposed in the temperature range 178.09°C -887.21°C, indicating that complex decomposes with to loss of C16H12N2O2 molecules .The total weight loss of this step is 85.94% which is in accordance with the calculated weight loss 86.78%.
TG fig. 2g to 2i of all the Co(II) complexes was carried out in N2 atmosphere with flow rate 50ml/min. and the heating rate was 8ºC min−1. The complex [C14H12CoN2O4] shows two decomposition steps. The first step (199.93°C – 202.94°C) and the second step (383.33°C -433.38°C) are accompanied by mass loss 59.16 % and 17.71 %, respectively. These first steps may be due to the decomposition of two phenols and one nitrogen molecules. The second step may be due to the loss of -C2H2 molecule. All the observed weight loss was found to be equal to theoretical weight loss. The complex was converted to metallic oxide as final residue.29 TG fig. 2h of C18H12CoN2O2 showed two decomposition steps in the temperature range 112.77°C to 168.55°C , with 13.24% (14.15%) weight loss, which was attributed to the loss of NO,1/2O2 moieties. In the second step the percentage weight loss 56.01 % (57.03%) weight loss was observed to loss of C12H11N group of the complex.
The mixed ligand complex of Co(II) shows two decomposition steps one is for loss of Hydrogen gas and two oxygen molecule from the complex.30 which were removed by purge gas nitrogen having flow rate 50ml/min. Then the second step was attributed to loss of [C7H7NO2] molecules, and the formation of CoO residue.
![Figure 2: Thermogravimetric analysis of the metal complexes, (a) to (i) in the temperature range from 40 ºC to 1000ºC (a) [C14H12 CuN2O4 ], (b) [C18H12CuN2O2], (c) [C16H12CuN2O2] , (d) [C14H12N2NiO4], (e) [C18H12NiN2O2 ], (f) [C16H12NiN2O2], (g) [C14H12CoN2O4 ], (h) [C18H12CoN2O2 ], (i) [C16H12CoN2O2].](http://www.orientjchem.org/wp-content/uploads/2016/12/Vol32No6_Nano_KOLH_fig2-150x150.jpg) |
Figure 2: Thermogravimetric analysis of the metal complexes, (a) to (i) in the temperature range from 40 ºC to 1000ºC (a) [C14H12 CuN2O4 ], (b) [C18H12CuN2O2], (c) [C16H12CuN2O2] , (d) [C14H12N2NiO4], (e) [C18H12NiN2O2 ], (f) [C16H12NiN2O2], (g) [C14H12CoN2O4 ], (h) [C18H12CoN2O2 ], (i) [C16H12CoN2O2]. |
X-ray diffraction analysis
The structures of complexes were confirmed by powder XRD recorded at 2θ and 20-80º. All the mixed ligand Cu (II), Ni (II) and Co (II) metal ion complexes of show broad and intense peak. The line broadening of the crystalline diffraction peaks in mixed ligands complexes of Co(II), Ni(II) and Cu(II) complexes shows higher crystallinity and fine nanoparticles. The XRD patterns of the mixed ligand complexes are shown in Fig. 3
![Figure 3: Powder XRD pattern for nanocrystalline mixed ligand complexes of a) [C16H12NiN2O2], b) [C16H12CuN2O2], c) [C16H12CoN2O2].](http://www.orientjchem.org/wp-content/uploads/2016/12/Vol32No6_Nano_KOLH_fig3-150x150.jpg) |
Figure 3: Powder XRD pattern for nanocrystalline mixed ligand complexes of a) [C16H12NiN2O2], b) [C16H12CuN2O2], c) [C16H12CoN2O2]. |
The unit cell parameters were obtained by using Powder -X software methods. The complexes of mixed ligands of the Cobalt, Nickel and Copper shows Monoclinic (P), Orthorhombic (P)31 and monoclinic (I) respectively. [JCPDS: 76-0476, JCPDS 43-1973). The diffraction data has been summarized in Table 4.
Table 4: Crystal data of the mixed ligands complexes of Co(II), Ni(II) and Cu(II).
| Chemical formula | [C16H12CoN2O2] | [C16H12NiN2O2] | [C16H12CuN2O2], |
| Formula weight | 339.21 | 338.97 | 343.83 |
| Crystal color | Chocolate Brown | Dark Olive Green | Green |
| Temperature (K) | 298 | 298 | 298 |
| Wavelength (Å) 0 | 1.54050 | 1.54051 | 1.540598 |
| Radiation | Cu Kα | Cu Kα | Cu Kα |
| Crystal system | Monoclinic | Orthorhombic | Monoclinic(I) |
| Space group | P21/a (14) | Pbna(60) | P21/n(14) |
| A | 1.4276 | 0.7806 | 0.7515 |
| C | 0.9396 | 1.2891 | 0.3915 |
| Z | 4 | 8 | 4 |
| a (Å) | 13.819(2) | 11.883(3) | 13.185(7) |
| b (Å) | 9.68(2) | 15.222(4) | 17.546(8) |
| c (Å) | 9.095(5) | 19.623(6) | 6.871(3) |
| α | 90.00 | 90 | 90.00 |
| β ( ) | 105.83 | 90 | 107.01 |
| γ () | 120.00 | 90 | 120.00 |
| a/b | 0.7449 | 0.7757 | 0.7388 |
| c/b | 0.5245 | 0.6056 | 0.3916 |
| Volume (Å3) | 1170.48 | 3549.47 | 1520.03 |
| Dx (g/cm3) | 1.891 | 1.509 | 1.738 |
Scanning Electron Microscopy and Electron dispersive Spectroscopy
The complexes were scanned by Scanning Electron Microscope is the good agreements for formation of the nanocrystalline complexes with best morphology. The SEM micrographs of these complexes were taken in the scale of 1μm, 5 μm, and 10 μm for each nanocrystalline mixed ligands metal complexes. The scanning electron micrograph of the complexes shown in Fig. 4 The complex [C18H12CuN2O2 ] shows the shape like nanoflowers,32 the Co(II) mixed ligand complex shows nanofoil shaped rod with internal hexagonal array.22 The Ni (II) mixed ligand complex of N, O donors shows the leaf like morphology. All the mixed ligand complexes have grain size less than 10 nm. The average size of these complexes were calculated by using 2θ, FWHM, and Cu-Kα radiation of wavelength 1·54058 Å from powder XRD with grain size measurement programmers software. The nanocrystalline sizes of mixed ligands complexes of Cu (II), Ni (II) and Co (II) are 7.90488nm; 6.83833nm and 7.31095nm respectively. The decrease in the size indicates the increased surface to volume ratio increases. The SEM study shows good agreement for the formation of nanocrystalline complexes and also supporting tool for the powder XRD. Therefore these complexes can be used as the catalysts in organic transformations.
 |
Figure 4: Scanning Electron Micrographs of Mixed ligand complexes taken on the scale of 1μ, 5μ, 10μ as a, b, c) SEM image of Co(II) mixed ligands complexes, d, e, f) SEM image of Ni(II) mixed ligands complexes , g, h, i) SEM image of Cu(II) mixed ligands complexes. Click here to View figure |
The Electron dispersive spectra of mixed ligand metal complexes were taken. The sample was coated on the Platinum plates at room temperature. The Electron dispersive spectroscopic studies of mixed ligand metal complexes show compositions of the nanostructured complexes. The EDS analysis data matched with C, H, and N elemental analysis result. The percentage of oxygen in all complexes analyzed by EDS was found to be high due to the oxygen adsorbed on the complexes .The complexes have nanostructures with array of different shapes in which array, the oxygen get adsorbed and percentage of oxygen get elevated. The EDS spectrum is shown in Fig 5.
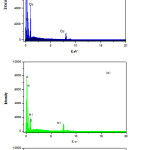 |
Figure 5: Electron Dispersive spectrum of nanocrystalline mixed ligand complexes of a) Co (II) , b) Ni(II) and c) Cu(II) . |
The data of elemental analysis are shown in Table 1. and EDS were as calculated (found) for the complex are [C16H12CuN2O2] C 55.89%(55.71%),N 8.15%(8.04%), O 13.16% (17.77%) ,Cu 18. 48%(18.40%). The complex [C16H12NiN2O3] contain was C 55.69 %(55.56%), N 8.16%(8.13%), O 14.16% (17.02%), Ni 17.32%(17.15%) and the mixed ligands complex of Cobalt as [C16H12CoN2O3] C 56.65%(56.66%),N 8.26%(7.64),O 14.15 (16.98), Co 17.37% (17.22%).The possible 3D-chemical structures homoleptic [ML1],[ML2] and heteroleptic [ML1L2] complexes were shown in fig. 6
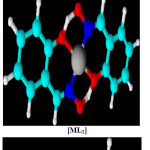 |
Figure 6: Suggested chemical bonding and 3D-structure for complexes. |
Antimicrobial activity of Complexes and Ligands
The result of antimicrobial activity of the synthesized complexes and ligands photo are shown in Fig.7.
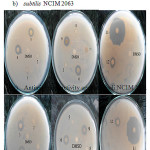 |
Figure 7: Photograph of Biological activity of Ll, L2 and their metal complexes a) Antimicrobial activity against Bacillus subtilis NCIM 2063 |
The complexes 3: [C16H12CuN2O2], 4:[C14H12N2NiO4],6:[C16H12NiN2O2],9:[C16H12CoN2O2] showed antimicrobial activity against both B. subtilis NCIM 2063 and E.coli NCIM 2341. All other complex is inactive against test organism. Both ligands L1: [C9H7NO] and L2: [C7H7NO2] showed antimicrobial effect. The L1:[C9H7NO] shown larger zone than L2 : [C7H7NO2] 33. In case of heteroleptic complexes of Cu(II), Co(II) and Ni(II) shows high antimicrobial activity as compared to the homoleptic complexes. The antimicrobial activity of all the Ligands and complexes were summarized in Table 5.
Table 5: Antimicrobial activity measured in Zone of inhibitions
| Sr. No. | Complex | Zone of inhibition (mm) for Bacillus subtilis | Zone of inhibition (mm) for E.coli |
| L1 | [C9H7NO] |
30 |
32 |
| L2 | [C7H7NO2] |
15 |
15 |
| 1 | [C14H12 CuN2O4 ] |
00 |
00 |
| 2 | [C18H12CuN2O2 ] |
00 |
00 |
| 3 | [C16H12CuN2O2] |
15 |
16 |
| 4 | [C14H12N2NiO4 ] |
15 |
16 |
| 5 | [C18H12NiN2O2 ] |
00 |
00 |
| 6 | [C16H12NiN2O2] |
17 |
15 |
| 7 | [C14H12CoN2O4 ] |
00 |
00 |
| 8 | [C18H12CoN2O2 ] |
00 |
00 |
| 9 | [C16H12CoN2O2] |
16 |
15 |
| 10 | [C9H7NO] |
00 |
00 |
Conclusion
The mixed ligands complexes of two N, O donor ligands were successfully synthesized and characterized by different spectral techniques and micro analytical methods. The synthesized mixed ligand complexes were nanocrystalline in nature with good morphology. All complexes have shown non-electrolytic behavior as observed from the conductance measurement. The powder XRD, SEM gives the result of formation of crystalline fine nanoparticles. The nanocrystalline mixed ligand complexes has the less than 10nm therefore these complexes may be used as catalyst in synthetic organic chemistry for functional group transformation. The nanocrystalline heteroleptic complexes show higher antimicrobial activity than other complexes. Among complexes [C16H12CuN2O2], [C14H12N2NiO4 ], [C16H12NiN2O2], [C16H12CoN2O2] have shown antimicrobial activity. Both ligands showed positive results. But activity of L1: [C9H7NO] was nearly double than L2 : [C7H7NO2].
Acknowledgments
The authors are very grateful to management of Ahmednagar Jilha Maratha Vidya Prasarak Samaj’s, Principal Dr.B.H.Zaware, New Arts Commerce and Science College Ahmednagar for providing laboratory facilities.
References
- Milios CJ, Stamatatos TC, Perlepes SP.,Polyhedron, 2006, 25, 134-194.
CrossRef - Babu MSS, Krishna PG, Reddy KH, Philip GH, Indian J. Chem., 2008, 47,1661–1665.
- R. Prasad , M. Mathur, J. Serbian Chem. Soc., 2002, 67, 825–832.
CrossRef - B. Khera, A. K. Sharma, N. K. Kaushik, Polyhedron, 1983, 2,1177–1180.
CrossRef - M. Kurtoglu, , J. Serbian Chem. Soc., 2010, 75, 1231–1239.
CrossRef - Raptopoulou CP, Sanakis Y, Boudalis AK, Psycharis V. Polyhedron, 2005, 24, 711–721.
CrossRef - Aggarwal RC, Singh NK, Singh RP, , Synth. React. Inorg. Met. Chem., 1984, 14,637–650.
CrossRef - Patel KD,Patel HS, A.J.of chem.2013,http://dx.doi.org/10.1016/j.arabjc.2013.03.019
CrossRef - Masalovich MS, Ardasheva LP, Shagisultanova G, Zh. Neorg. Khim., 2006, 51,1591–1596.
- Thakkar N V., Thakkar JR., Synth. React. Inorg. Met. Chem., 2000, 30, 1871–1887.
CrossRef - Prasad RV, Thakkar NV., J. Mol. Catal., 1994, 92, 9–20.
CrossRef - Angelusiu MV, Barbuceanu SF, Draghici C, Almajan GL. Almajan, Eur. J. Med. Chem., 2010, 45, 2055–2062.
CrossRef - Hartophylles M., Zeitschrift fur anorganische und allgemeine Chemie, 1995, 621,645–653.
CrossRef - Sallam SA., Transit. Met. Chem., 2006, 31, 46–55.
CrossRef - Alemi AA, Shaabani B, B. Shaabani, Acta Chim. Slov., 2000,47, 363–369.
- Baran EJ., Zeitschrift fur Naturforsch. – Sect. B J. Chem. Sci., 2005,60, 663–666.
- Cupertino D, Mcpartlin M, Zissimos AM, 2001, 20, 3239–3247
- Downing RS, Urbach FL, J. Am. Chem. Soc., 1969, 91,5977–5983.
CrossRef - Patil SS, Thakur GA, Shaikh MM., ISRN Pharm.,2011, 2011, 1–6.
- Cristóvão B., J. Serbian Chem. Soc., 2011,76,1639–1648.
CrossRef - Jain SK, Garg BS, Yudhvir K, 1987, 77, 73–77.
- Ibrahim KM, Bekheit MM., Transit. Met. Chem., 1988, 232, 230–232.
CrossRef - Li H, Li Y., Nanoscale, 2009,1, 128–132.
CrossRef - Firenze UDI, Inorg. Chem., 1966, 5, 1960–1963.
CrossRef - Raman N, Ravichandran S, Thangaraja C, J. Chem. Sci., 2004,116, 215–219.
CrossRef - Turkoglu O, Soylak M, Belenli I., Collect. Czechoslov. Chem. Commun., 2003, 68,1233–1242.
CrossRef - Mohamed GG, El-wahab ZHA., 2003, 73, 347–359.
- Mashaly MM, Abd-Elwahab ZH, Faheim AA., J. Chinese Chem. Soc., 2004, 51, 901–915.
CrossRef - Abdallah SM, Zayed MA, Mohamed GG, Arab. J. Chem., 2010, 3, 103–113.
CrossRef - Nayak SC, Das PK, Sahoo KK., Chem. Pap., 2003, 57, 91–96.
- Patel KS, Patel JC, Dholariya HR, Patel VK, Patel KD, 2012, 2012, 49–59.
- Liu B, Zhu S, Zhang W, Chen C, Zhou Q, Q. Zhou, 2007, entry 10, 5834–5835.
- Shao Q, Wang T, Wang X, Chen Y. Chen, Front. Optoelectron. China, 2011, 4, 195–198.
CrossRef - Bagihalli GB, Avaji PG, Patil SA, Badami PS, Eur. J. Med. Chem., 2008, 43, 2639–2649.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









