Synthesis and Characterisation of Biologically Potent Novel Chalcone Moieties
Mahammadali Khanusiya* and Z. M Gadhawala
Department of Chemistry, The HNSB Ltd Science College, Himatnagar, Gujarat-India.
Corresponding author Email: khanusiya.mali@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/320244
Article Received on :
Article Accepted on :
Article Published : 07 May 2016
As displaying a dominant biological interest of some amino chalcone derivatives which were synthesized by claisen-schmidt condensation reaction of amino acetophenone with aromatic aldehyde in presence of sodium hydroxide. These chalcones were screened for antifungal activity against candida albicans strain and also for antibacterial activity against staphylococcus epidermidis (G positive) and pseudomonas aeruginosa (G negative) strain by NCCLS method. The synthesized compounds were characterized by means of their FT-IR and 1HNMR spectral study.[14]
KEYWORDS:Amino chalcone; Antifungal; Antibacterial
Download this article as:| Copy the following to cite this article: Khanusiya M, Gadhawala Z. M. Synthesis and Characterisation of Biologically Potent Novel Chalcone Moieties. Orient J Chem 2016;32(2) |
| Copy the following to cite this URL: Khanusiya M, Gadhawala Z. M. Synthesis and Characterisation of Biologically Potent Novel Chalcone Moieties. Orient J Chem 2016;32(2). Available from: http://www.orientjchem.org/?p=15896 |
Introduction
Chalcones are pharmacologically valuable moieties possessing 1,3diphenyl prop-2-ene-1-one (-CH=CH-CO-) as a core structure in which two aromatic rings are linked by first and third carbon of a α,β- unsaturated carbonyl skeleton. A number of chalcone derivatives have demonstrated wide spectrum of pharmacological activities which has drawn the attention of medicinal chemist and pharmacologists. Due to the extended conjugation, the complete delocalisation of π electrons on both the benzene rings makes whether from bioactivity aspect.
Isolation of Chalcone derivatives from nature requires a long and a far complicated procedure and comparable yield does not obtained. Chalcones and their derivatives are an interesting target class of compound which are extensively investigated due to their broad spectrum of various therapeutic activity such as antimicrobial[1], anti-inflammatory[2], antiulcerative[3], antiviral[4], antifungal[5], antimalarial[6,7], and anticancer[8]. Furthermore, chalcones are also known as the key intermediate in the synthesis of various biologically active heterocyclic compounds. In order to synthesize new therapeutic agents, this report illustrated the some novel amino chalcone derivatives and screening their activity against candila albicans fungi, gram positive and gram negative bacterial species.
Material And Method
General information
The all starting materials and solvents were purchased from sigma-Aldrich and SD Fine and used without further purification. Melting points were determined by conventional method and then by electro capillary apparatus and are uncorrected. All the synthesized compounds were inspected by thin layer chromatography on silica gel (E-Merck) and the spots were identified by UV lamp.IR spectra and proton NMR spectra in DMSO at 500 MHz were recorded at CSMCRI Bhavnagar.
General procedure of synthesis of chalcone
The synthesis of chalcone derivatives was conducted according to the procedure reported in the reference [10-12]. Acetophenone derivative (2.5milimole) and substituted benzaldehydes (2.5milimole) were dissolved in 30 ml methanol. To the solution, 10 ml NaOH(20%) solution was added drop wise and reaction mixture was stirred for 1-2 hour at room temperature by magnetic stirrer and kept for overnight. Subsequently, it was poured in ice water and neutralized. The solid precipitates was filtered off and recrystallized from methanol or ethyl acetate.
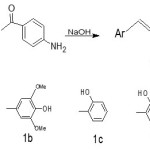 |
Scheme 1 Click here to View scheme |
Result and Discussion
1a; (2E)-1-(4-aminophenyl)-3-(4-hydroxy-3-methoxyphenyl)prop-2-en-1-one
Yellow solid, Yield 58.9 %. M.P 105-107 ᵒC, Rf 0.71
FT-IR (ν, cm-1): 3395(-OH), 3330, 3225 (-NH2), 3063(aromatic C-H), 1649 (>C=O), 1590(-HC=CH-), 1277, 1305 (C-N str)
1HNMR (500 MHz CDCl3, Me4Si): 3.50 ( br, s, -NH2), 3.85(s, -OCH3), 6.027(s,1H, H2), 6.62( d, 1Hα), 7.66 ( d, 1Hβ),.
1b; (2E)-1-(4-aminophenyl)-3-(4-hydroxy-3,5-dimethoxyphenyl)prop-2-en-1-one
Yellow solid, Yield 57.5 % . M.P 98-100 ᵒC , Rf 0.66
FT-IR (ν, cm-1): 3396(-OH), 3331, 3225(-NH2), 3064(aromatic C-H), 1648 (>C=O), 1590 (-HC=CH-), 1276, 1304 (C-N str)
1HNMR (500 MHz DMSO, Me4Si): 3.49( br, s, -NH2), 3.71(s, -OCH3), 3.82(s, -OCH3), 6.028(s,1H, H1,H6), 6.58( d, 1Hα), 7.68 ( d, 1Hβ).
1c: (2E)-1-(4-aminophenyl)-3-(2-hydroxyphenyl)prop-2-en-1-one
Pale Yellow solid, Yield 62.5% . M.P 110-112 ᵒC ,Rf 0.68
FT-IR (ν, cm-1): 3570 (-OH), 3338, 3328 (-NH2), 3050(aromatic C-H), 1676(>C=O), 1595(-HC=CH-), 1270,1364 (C-N str)
1HNMR (500 MHz CDCl3, Me4Si): 3.47 ( br, s, -NH2),6.57( d, 1Hα), 7.51 ( d, 1Hβ), 7.013 ( m, H2), 7.72 (m, H3,H4).
1d: (2E)-1-(4-aminophenyl)-3-(2,4-dihydroxyphenyl)prop-2-en-1-one
Yellow solid, Yield 87.0% . M.P 98-100 ᵒC ,Rf 0.72
FT-IR (ν, cm-1): 3680 (-OH), 3652 (-OH), 3370, 3325(-NH2), 3062(aromatic C-H), 1673 (>C=O), 1596(-HC=CH-), 1270,1364 (C-N str)
1HNMR (500 MHz CDCl3, Me4Si): 3.48( br, s, -NH2),6.32(s,1H, H2), 6.55( d, 1Hα), 7.48 ( d, 1Hβ), 7.55 (d, 1H, H4), 7.67(d,1H, H5).
1e: (2E)-1-(4-aminophenyl)-3-(2-hydroxynaphthalen-1-yl) prop-2-en-1-one
Brick Red solid, Yield 73.6% .M.P 182-185 ᵒC, Rf 0.53
FT-IR (ν, cm-1): 3652(-OH), 3322, 3335(-NH2), 3044(aromatic C-H), 1668 (>C=O), 1587(-HC=CH-), 1267,1349 (C-N str)
1HNMR (500 MHz CDCl3, Me4Si): 3.49( br, s, -NH2),6.58( d, 1Hα), 7.68 ( d, 1Hβ), 7.42 (m, C-H,), 7.85(d,1H, H3),8.05(d,1H, H2).
Antibacterial Study
Gram Positive microorganism [Staphylococcus epidermidis]
Table-1: Percentage cell inhibition by compounds agains Staphylococcus epidermidis strain.
| Conc (μg/ml) | Log Conc. |
1a |
1b |
1c |
1d |
1e |
STD |
|
0.01 |
-2.29 |
1.023 |
12.23 |
1.08 |
11.02 |
7.16 |
10.12 |
|
0.02 |
-1.82 |
1.02 |
14.11 |
5.22 |
10.22 |
8.02 |
11.89 |
|
0.05 |
-1.34 |
4.05 |
17.02 |
7.13 |
14.74 |
9.11 |
13.98 |
|
0.14 |
-0.86 |
5.11 |
21.26 |
22.41 |
17.23 |
13.21 |
18.03 |
|
0.41 |
-0.39 |
11.23 |
26.17 |
30.27 |
28.21 |
15.42 |
28.49 |
|
1.23 |
0.09 |
14.52 |
32.20 |
32.36 |
27.62 |
18.85 |
33.26 |
|
3.70 |
0.57 |
25.34 |
39.19 |
42.87 |
40.33 |
25.63 |
42.13 |
|
11.11 |
1.05 |
29.78 |
52.36 |
48.16 |
63.11 |
40.17 |
49.65 |
|
33.33 |
1.52 |
37.41 |
68.23 |
49.20 |
74.42 |
67.32 |
70.32 |
|
100.00 |
2.00 |
67.28 |
74.12 |
54.14 |
93.12 |
87.24 |
84.97 |
|
IC50 μg/ml |
7.69 |
5.67 |
0.30 |
7.60 |
21.61 |
7.625 |
|
|
R2 |
0.9227 |
0.9760 |
0.9632 |
0.9773 |
0.9923 |
0.9507 |
|
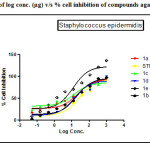 |
Figure 1: Plot of log conc. (µg) v/s % cell inhibition of compounds against Staphylococcus epidermidis. Click here to View figure |
Gram Negative bacteria [Pseudomonas aeruginosa]
Table 2: Percentage cell inhibition by compounds against Pseudomonas aeruginosa strain.
| Conc(μg/ml) | Log Con. |
1a |
1b |
1c |
1d |
1e |
STD |
|
0.01 |
-2.29 |
7.43 |
14.32 |
8.10 |
1.23 |
4.30 |
8.00
|
|
0.02 |
-1.82 |
10.22 |
15.26 |
12.42 |
2.52 |
7.12 |
12.94 |
|
0.05 |
-1.34 |
11.14 |
15.79 |
15.04 |
6.21 |
11.25 |
9.67 |
|
0.14 |
-0.86 |
17.16 |
20.64 |
22.16 |
5.11 |
13.02 |
13.41 |
|
0.41 |
-0.39 |
21.03 |
29.71 |
27.65 |
9.21 |
16.27 |
18.87 |
|
1.23 |
0.09 |
22.17 |
32.27 |
35.44 |
16.23 |
19.15 |
23.42 |
|
3.70 |
0.57 |
30.13 |
42.24 |
39.87 |
35.21 |
22.27 |
26.51 |
|
11.11 |
1.05 |
42.16 |
48.24 |
45.31 |
39.47 |
35.65 |
29.04 |
|
33.33 |
1.52 |
58.23 |
64.64 |
52.55 |
47.11 |
47.68 |
52.67 |
|
100.00 |
2.00 |
64.12 |
83.12 |
81.65 |
54.20 |
77.24 |
76.19 |
|
IC50 μg/ml |
7.908 |
3.795 |
10.30 |
2.795 |
34.94 |
9.871 |
|
|
R2 |
0.9737 |
0.9384 |
0.8675 |
0.9863 |
0.9643 |
0.9473 |
|
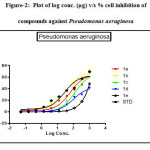 |
Figure 2: Plot of log conc. (µg) v/s % cell inhibition of compounds against Pseudomonas aeruginosa Click here to View figure |
Antifungal activity
Table 3: Percentage growth inhibition of compounds against fungal strain and Fluconazole (Std.)
|
Conc(μg/ml) |
LogConc |
1a |
1b |
1c |
1d |
1e |
Std |
|
0.05 |
-1.29 |
-0.02 |
0.35 |
0.36 |
-0.35 |
0.22 |
5.68 |
|
0.15 |
-0.82 |
0.032 |
2.36 |
0.89 |
2.35 |
0.156 |
10.64 |
|
0.46 |
-0.34 |
6.235 |
6.58 |
2.37 |
4.79 |
2.36 |
14.47 |
|
1.37 |
0.14 |
11.35 |
13.26 |
5.46 |
9.63 |
7.86 |
18.63 |
|
4.12 |
0.61 |
24.65 |
17.89 |
18.65 |
18.67 |
14.62 |
32.87 |
|
12.35 |
1.09 |
34.68 |
44.65 |
21.34 |
25.63 |
21.35 |
49.58 |
|
37.04 |
1.57 |
51.34 |
58.96 |
29.62 |
37.48 |
27.65 |
65.65 |
|
111.11 |
2.05 |
58.36 |
67.25 |
31.28 |
46.22 |
34.52 |
68.61 |
|
333.33 |
2.52 |
68.34 |
76.38 |
48.96 |
61.23 |
48.75 |
87.09 |
|
1000.00 |
3.00 |
77.35 |
84.36 |
50.30 |
71.22 |
66.22 |
99.08 |
|
IC50 μg/ml |
13.12 |
12.69 |
19.02 |
32.29 |
74.37 |
14.39 |
|
|
R2 |
0.9809 |
0.9895 |
0.9358 |
0.9680 |
0.9394 |
0.9685 |
|
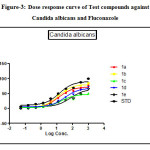 |
Figure 3: Dose response curve of Test compounds against Candida albicans and Fluconazole Click here to View figure |
The antibacterial activity performed by antimicrobial susceptibility tests, NCCLS 1993, Approved standard: M2-A5. Chemically synthesized compounds were taken for antibacterial activity. P.aeruginosa (Gram negative), S.epidermidis (Gram positive) strains were used for the antibacterial activity. Three-fold serial dilutions of the compounds (150µl) of each sample were made in sterile broth (nutrient broth). The specified amount of test organisms (50µl) was added to each dilution to give a final volume of 200µl. After incubation at 37 °C for 18–24 h the plates were examined for growth of the organisms. Absorbance was read in a plate reader. The figures showed the plot of log concentration Vs % cell inhibition of compound against tested organisms. Test compounds show the dose-effect co-relation with maximum linearity in almost all cases within 0.8321 to 0.9863. The data analysis was accomplished using Graph Pad Prism version 5.00, Graph Pad Software Inc., San Diego California USA. IC50 values were obtained from regression lines with coefficient factors between R2 = 0.52 and 0.99. Absorbance at 595nm between reading taken before and after incubation of the plates.
Results of gram positive organism inhibition study indicate followings compounds were found to be lid among others when it was compare IC50 value obtained by Streptomycin (7.625 µM/ml) with standard drug inhibitory value.
1c (IC50 value; 0.3086 µM/ml, R2= 0.9632)> 1b (IC50 value; 5.303µM/ml, R2= 0.8821) >1d>1a
Results for gram negative microorganism, most effective compounds were arranged in following order when it was compare IC50 value obtained by Streptomycin (9.871 µM/ml) with standard drug inhibitory value:
1d (IC50 value; 2.795 µM/ml, R2= 0.9863)>1b (IC50 value; 3.795 µM/ml, R2= 0.9384)>1a(IC50 value; 7.908 µM/ml, R2= 0.9737) >1c(IC50 value; 9.871 µM/ml, R2= 0.9636)
Fluconazole and test compounds were tested against Candida albicansin dose dependent manner. Table showed that the high IC50 value of 1e was found against Candida albicans having (IC50: 74.37µg/ml). Comparative lower IC50 value was found with compounds 1b against candida albicans(IC50: 12.6µg/ml).The figure showed the plot of log concentration vs. % cell inhibition of test compounds against Candida albicans. Test compounds show the dose-effect co-relation with maximum linearity (R2value) with value being 0.9895 and comparatively lower linearity in case of Candida albicans at value being 0.9358. Activity order of compounds can be summarized as :1b>1a>1c>1d.
Conclusion
From above results we can conclude that compounds 1a, 1b and 1c are more effective in gram positive organism i.e. Staphylococcus epidermidis, While compounds like 1a, 1b and 1d are more effective against gram negative organism i.e. Pseudomonas aeruginosa.
Moreover, 1a and 1b both show good antifungal activity on Candida albicans strain, while 1c and 1d having comparable activity and compound 1e is less effective.
Acknowledgement
The author M M Khanusiya gratefully acknowledges CSIR, New Delhi for MANF. The authors are also expressing their sincere thanks to The HNSB Ltd Science College for performing research work and faculty of pharmacy, DDU Nadiad for antimicrobial tests.
References
- Prasad, Y.R; Kumar, P.R; Deepti, C.A; Ramna, M.V; E-Journal of chem, 2006, 3(3), 236-241
CrossRef - Heish, H.K; Lee, T.H; Wang, J.J; Lin, C.N; Phasm. Res. 1998, 15(1), 39-46
- Murakami, S; Muramatsu, M; Aihara, H; Otomo, S; Biochem. pharma, 1991,42(7), 1447-1451
- Wu, J.H; Wang, X.H; Yi, YH; Lee, K.H; Bioorg. Med.Chem.Lett, 2003, 13(10), 1813-1815.
CrossRef - Lopaz, S.N; Castle, M.V; Jacchino, S.A; Dominguez, J.N; Lobo, G; Cortsey, J.C; Ribas, J.C; Devia, C; Rodrigues, A.M; Enriz, R.D; Bioorg. Med.Chem. 2001, 9(8), 1999-2004.
CrossRef - Leon, C; Gut, J; Rosenthal, P.J; J.Med.Chem.2005, 48, 3654.
CrossRef - Ram, V; Saxena, A; Srivastava, A; Bioorg. Medicinal chemistry lett.2000,10,2159
CrossRef - Liu, M; Wilairat, P; Crajt, S; Bioorg. Med. Chem. 2003,11,2729.
CrossRef - Kumar, S.K; Hager, E; Pettit, C; Gurulingappa, H; Devidson,N.E; Khan, S.R; J. Med. Chem. 2003,46,2813-2815.
CrossRef - Suvito, H; Jumina; Mustofa; Mutuzahroh , Ni; Nyoman, Ni; Der Pharma Chemica. 2015, 7(3), 89-94.
- Ngameni, B; Kuete, V; Ambasa, P; Justine, K; Marlyse, M; Medicinal Chem 2013, 3; 233-237.
- Abdula A; European Journal of Chemistry 2013, 4(2), 207-210J.
CrossRef - Pngaew, R; Saekee, A; Mandi, P; Nantasenamat, C; Prachyasittikul, S; Ruchirawat ,S; Prachyasittikul, V; European Journal of Chemistry, 2014,85,65-76.
CrossRef - Khanusiya, M.M; Gadhawala Z.M; National Seminar on ‘Frontier Areas In Chemical Sciences’, 2016, 42.

This work is licensed under a Creative Commons Attribution 4.0 International License.









