InhA inhibitors as potential antitubercular agents
Hinna Hamid
Department of Chemistry, Faculty of Science, JamiaHamdard, Hamdard Nagar, New Delhi 110062, India
DOI : http://dx.doi.org/10.13005/ojc/320106
Article Received on :
Article Accepted on :
Article Published : 12 Mar 2016
Infectious diseases pose a potential threat and are responsible for killing more people globally than any other single cause. For six decades, antibiotics have been a bulwark against bacterial infectious diseases. This bulwark is failing due to the appearance of resistant bacterial strains. The biosynthesis pathway of fatty acids in bacteria represents validated target for antibacterial drug discovery, although it is still relatively unexplored. Bacterial FAS-II is essential for survival of bacterial cells, furthermore as itis distinct from FAS-I pathway in mammals it constitutes a rational beginning point for the design of novel inhibitors of FAS-II enzyme.Tuberculosis (TB) is a potentially serious infectious disease. Several strategies that have been adopted for the development of newtherapeutic agents against the strains of tuberculosis which are drug resistant involve not only the design and synthesis of compounds against earlier known targetsbut also the identification and inhibition of novel drug targets. Isoniazid (INH), the first-line agent used for prevention and treatment of tuberculosis targets the enoylreductase (InhA) enzyme in fatty acid biosynthesis pathway of the mycobacteria. KatG, is the enzyme which activates isoniazid and mutations in this enzyme lead to majority of INH-resistant clinical strains. Thus only those compounds will work against most of the INH resistant strains of mycobacteria which do not require activation of KatGfor InhA inhibition.
KEYWORDS:InhA; antibacterial; FAS II; rational design; FabI inhibitors; enoyl ACP reductase; FAS
Download this article as:| Copy the following to cite this article: Hamid H. InhA inhibitors as potential antitubercular agents. Orient J Chem 2016;32(1). |
| Copy the following to cite this URL: Hamid H. InhA inhibitors as potential antitubercular agents. Orient J Chem 2016;32(1). Available from: http://www.orientjchem.org/?p=14888 |
Introduction
Tuberculosis (TB) is a potentially serious infectious disease. It has been reported that one third population of the world is affected with Tuberculosis and 9.6 million people suffered from Tuberculosis in 2014.In 2014 almost 9,421 people suffered from Tuberculosis (a rate of 2.96 cases per 100,000 persons) in the United States [1]. Mycobacterium tuberculosis (MTB) is a leading killer in patients suffering from AIDS and because of the advent of multidrug-resistant strains of MTB (MDRTB)) existing control measures are severely hindered[2-4]. The spread of this major disease is being prevented through chemotherapy as no effective vaccine is available. Isoniazid (INH) is one of the most potent and widely accepted drugs for treating tuberculosis and constitutes the current front line treatment regimen. Its mechanism of action includes disrupting the integrity of the mycobacterial cell wall byimpedingmycolic acid biosynthesis [5]. Mycobacterium. tuberculosisstrains were found to develop resistance to it almost immediately following its clinical use and about 30% of the clinical isolates currently are INH resistant. The mechanism of its action remained unclear until 1994, although it was introduced in 1952. It was found out that due to a missense mutation of theinhAgene led to development of resistance to isoniazid[6] and thatinhAgene in the Mycobacterium tuberculosis coded for the FabI protein (InhA)[7]. Isoniazid requires a prior activation by the KatG catalase/peroxidase and thus acts as a pro-drug,which is the underlying cause for fast development of resistance to isoniazid [5, 8-10]. M. tuberculosisstrains which are catalase/peroxidase-deficient remain virulent [11]. Despite the fact that the mechanism of action of isoniazid is complicated it is evident that InhA, the enoylreductase (ENR) in the type II biosynthesis pathway offatty acids, is the target for isoniazid, [12-15], [6, 7, 16]. InhA was validated as a drug target for isoniazid byJacobs and co-workers by revealing that a mutation caused by change in temperature caused inactivation of InhA, which led to the same phenotypic response as administration of Isoniazid [26]. Inhibition ofInhA is affected by formation of an adduct between NAD(H) andINH after activation by KatG[10, 17-21] and compounds that could inhibit InhA without the requirement of activation by KatG, have remarkablepotential as novel drugs for fighting MDRTB [22, 23][9, 22, 24, 25].
Having known that for isoniazidtargetsInhAseveral approaches such as computational simulation [27, 28], selective optimization of side activities (SOSA)[29] and de novo design [30] have been utilized for rational design of novel inhibitors of InhA with sub-nanomolarpotency[31, 32].
Classes of Inhibitors
Three different classes of known inhibitors of ENR are based on isoniazid,diazaborines, and triclosan, the antibacterial agents that have shown to inhibit ENR. Two approaches, i.e., synthesis of several novel INH basedantimycobacterial compounds or INH-NAD adductshave been used to developisoniazid based molecules having better anti-mycobacterial potency. The other scaffolds identified by high-throughput screening (HTS) and exhibiting potent inhibition of ENR include piperazine, pyrrolidinecarboxamides, pyrazoles, imidazopiperidines, indoleformamidesarylamides and arylcarboxamides.
In this review an overview of the existing status of ongoing efforts to develop inhibitors for enoylreductase (FabI) involved in the catalysis of a critical step in the FAS II pathway has been provided.
Structural Generalizations
Although the known InhA inhibitor classes are structurally diverse, two generalizations arise. The inhibitors bind to the enzymein almost every case in presence of the oxidized and/or reduced cofactor, though isoniazid and the diazaborinesbind as covalent adducts of the cofactor. Also that the efficiency of inhibition is often related to ordering of the loop of the enzyme meant for substrate binding located in the vicinity of the active site, [33] when slow onset inhibitors interact with the enzyme. An initial rapid association of enzyme and inhibitor precedes the slow complex formation step. Slow onset enzyme inhibitors remain bound to their target enzymes even in case of low free drug concentration and thus have long residence times on the enzyme, prolonging their activity [34-36].
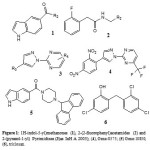 |
Figure 1 Click here to View figure |
In this review an overview of the existing status of ongoing efforts to develop inhibitors for enoylreductase (FabI) involved in the catalysis of a critical step in the FAS II pathway has been provided.
Staveskiet al., in (2001) reported synthesis of (1H-indol-5-yl) methanones (1), 2-(2-fluorophenyl) acetamides and 2-(pyrazol-1-yl) pyrimidines (3) as InhA inhibitors, affording (2) [R2 = 4-ClC6H4; n = 2] which showed 82% InhA inhibition at 40 µM [37] (Fig. 1). Kuoet al.,[32] used high throughput screening to
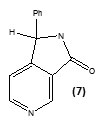
Staveskiet al., in (2001) reported synthesis of (1H-indol-5-yl) methanones (1), 2-(2-fluorophenyl) acetamides and 2-(pyrazol-1-yl) pyrimidines (3) as InhA inhibitors, affording (2) [R2 = 4-ClC6H4; n = 2] which showed 82% InhA inhibition at 40 µM [37] (Fig. 1). Kuoet al.,[32] used high throughput screening to identifypiperazine and pyrazolebased inhibitors ofInhA, the most effective compound (5) ((4-(9H-fluoren-9-yl)piperazin-1-yl)(indolin- 5-yl)methanone) was found to have an IC50 value for InhA of 0.16 µM and (4-(trifluoromethyl)-2-(4,5-dihydro-4-(2,4-dinitrophenyl)pyrazol-1-yl)pyrimidine) (4) exhibited an IC50 of 2.4 µM, respectively. Although the piperazine-based compound was found to be less potent in vitro than the pyrazole derivative itrepressed drug-resistant mycobacterium strains exhibiting an MIC99 value of 1–30 µM (Fig. 1).A sequence of ortho-metallation -electrophilic substitution was reported to be used as a critical step byBroussy, S. et al.,(2005)[38]for synthesis of 4-benzoylpyridine (7) scaffold. N-pyridyl alkylated analogue and 4-benzoylpyridine-3 carboxamide derivative which mimic the truncated Isoniazid–NAD adducts were present in a unique cyclized hemiamidal structure rather than the keto-amide open form. He et al., (2006) [39] used microarray parallel synthesis followed by high-throughput screening to report the discovery of various pyrrolidine carboxamides as a new class of inhibitors of ENR. On the basis of the preliminary SAR studies, a series of pyrrolidinecarboxamidederivatives were synthesized through an amide forming reaction by modifying the pyrrolidinecarboxamide frame work,in situ screening of these compounds led to discovery of more effective inhibitors based on pyrrolidinecarboxamide scaffold. Incidentally none of the compounds was found to exhibit any activity against the malaria parasite Plasmodium falciparum (pfENR) or ecENR(enoylreductase of E. coli). Rapid optimization of initial leads by structure-based iterative library approach led to development of inhibitors of InhA with nanomolar potency and compound (8) was found to be most active inhibitor exhibiting an IC50 of 140 nM. On evaluation of the pure enantiomers following the resolution of racemic mixtures it was found out that only one of the stereoisomersled to the inhibition showing an IC50 of 62 nM.
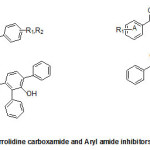 |
Figure 2 Click here to View figure |
A new class of arylamide based direct inhibitors of Inh A were discovered by Xinet al., (2007) using a high-throughput screening approach. These molecules were found to evade the mechanism of resistance to important antitubercular drugs likeIsoniazid and ethionamideas they do not require any mycobacterial enzymatic activation. Aseries of compounds were synthesized by diversifying the amine group but retaining the piperazinescaffoldusing SAR studies (Fig.2). More potent InhA inhibitors were discovered by in situ screening using an expedient microtiter library synthesis. Compound (9) exhibiting an IC50 of 90 nM[40] was found to exhibit most potent inhibition.Unique structural features of Triclosan, the broad spectrum antibacterial compound, have been used for the development of morediphenyl ethers derivatives with better inhibition of enoylreductase. Novel alkyl substituted diphenyl ether based InhA inhibitors have been developed by introducing various alkyl groups in place of chlorine atom on the phenol ring. A patent for diphenyl ether analogues as inhibitors of InhA has been claimed by Tongeet al., [41]. A library of alkyl substituted diphenyl ethers has been discovered to exhibit uncompetitive inhibiton of InhA. 8PP (10) the most potent compound, was reported to exhibit a K’i value of 1 n Magainst InhA and MIC99 values of 6-10 mM for both drug-sensitive and drug-resistant strains of MTB[31, 42]. In order to assess [43] the potential of imidazopiperidines as inhibitors of FAS II forM. tuberculosis growth as lead compounds all the synthesized compounds were evaluated for inhibition of InhA using triclosan (IC50 = 2.1 µM), as the reference and the compounds exhibiting effective inhibition were further subjected to whole M. tuberculosis cell assay.
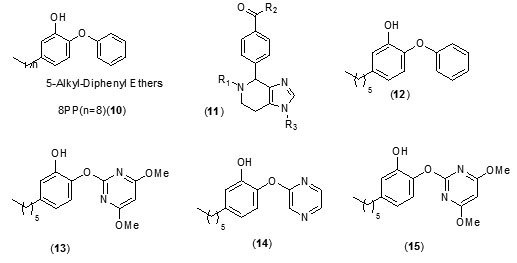
Preliminary observations showed that both R1 and R2 positions needed to be substituted for this scaffold to be active (11).
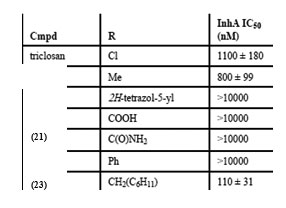
Compounds containing either the mono or dichloro benzyl substituent at the position R1 with R2 position having the electron donating para-methoxybenzylamine group (12, IC50= 0.24 µM) were found to be the most active. Removal of one chloride group was found to lead to decrease in activity (13, IC50 = 0.41 µM). SAR analysis of a group of hexyl substituted diaryl ethers with nitro-substituted phenyl ring or pyridine ring based InhA inhibitors reported by Endeet al.,[44]recommended that presence of a bulky group on the phenyl ring or phenyl ring with a nitrogen atom led to reduction of anti-InhA activity. 5-hexyl-2-phenoxyphenol (12) found to be a nanomolar inhibitor of InhA was also found to be effective against both sensitive as well as the isoniazid-resistant strains of M.tuberculosis showing an MIC90 of 1–2 µg/ mL. Although it displayed a significant in vitro activity, its ClogP value was over 5 and thus in order to reduce the lipophilicity and hence enhance the bioavailability, several B ring derivatives of (12) with either nitrogen containig heterocyles or nitro, amino,amide or piperazine substituted phenyl rings were designed and synthesized. Compounds (13), (14) and (15) showed MIC90 comparable with (12) andbetter ClogP. LeapFrog program was used to carry out CoMFA analysis of pyrrolidinecarboxamides which were reported as selective mtInhA inhibitors followed by subsequent de novo ligand design by Ashutoshet al.,[44]. The designed pyrrolidinecarboxamide derivatives showed better predicted activity in comparison to already reported molecules using the CoMFA model, suggesting that the proposed molecules could be more selective and effective against InhA. A tripeptide inhibitor with the sequence WYW was identified using a structure-based approach with the help of the crystal structure of Inh A with a known series of pyrrolidinecarboxamides. It was found have 100 fold more potency than the known inhibitors and could thus act as a useful lead molecule for further development of novel anti-TB compounds[30].For discovering novel and direct InhA inhibitors in silicoan effective approach was developed by Xiao-Yun Luet al., in 2009. Apharmacophore model was built employing bioactive conformation of pyrrolidinecarboxamide inhibitor bound to InhA. This model was effectively applied to screen the SPECS database in order to identify the biologically active conformations of pyrrolidinecarboxamide derivatives and align them; moreover a 3D-QSAR model which was statistically valid was also obtained. This led to selection and identification of 30 hit molecules as good leads from the database screening, which was followed by docking studies on molecules 16 and 17 to explore their interactions with ENR. Furthermore based on the generate dpharmacophore model the interactions between known pyrazole based inhibitors and ENR were analyzed[27].Using the approach of structure-based drug design several 5-substituted triclosan derivatives were synthesized in an attempt to develop diphenyl ethers with improved activity against InhA. Two groups of derivatives developed with enhanced potency against purified InhA had alkyl and aryl substituents. Most potentInhA inhibitor exhibiting IC50 of 21 nM displayed 50 timesbetter potency than triclosan. Structural attributes required for the inhibition were revealed by the study of four triclosan derivative bound X-ray crystal structures of ENR. Among the six derivatives selected and tested the most potent inhibitor (23) exhibited ten times enhancement of the anti-bactericidal activity in comparison with triclosan and was found to display an MIC of 4.7 μg mL-1 (13 μM). A subset of these triclosan analogs were also reported to be more potent against two strains of M. tuberculosisresistant to isoniazid, pointing to the importance of such structure-based design strategies for development of efficient drugs again sttuberculosis[45].
Ki of 22 pM. Analysis of theX-ray crystallographic structure of PT70 with ENR verified the fact that ordering of the loop for substrate-binding led to slow onset inhibition of the enzyme. The presence of the methyl group on B-ring of PT70 was found to exhibit crucial van der Waal’s interactions with important amino acid residues of the formerly disordered loop for substrate-binding and NAD cofactor. These results thus provided an insight into thestructural requirements for slow onset inhibition of ENR. It also pointed to the fact that PT70 had a significant potential to be effective against sensitive as well as drug-resistant strains of Mycobacterium tuberculosis[46]. Arylamides were found to be associated with high activity in ENR enzyme assay, but low antimycobacterial efficacy. In order to understand the structural activity relationship for improving the antimycobacterial activity, the dynamic behavior analysis of arylamide inhibitors and trans-2-hexadecenoyl-(N-acetylcysteamine)-thioester, a substrate was carried out by molecular dynamics (MD) simulation study. Arylamide inhibitors and the substrate were found to be positioned at the same site indicating that they acted as competitive inhibitors. The oxygen of the amide carbonyl was found to confer selectivity for InhA inhibition. Moreover, it was also found that this group was critical for hydrogen bond formation of thesearylamides with NADH and Tyr158 of InhA. It was thus realized that aryl ring substitution by hydrophilic groups enhanced theselectivity of these compounds to InhAand incorporation of more lipophilic groups into the substituent B resulted in the increase of their membrane permeability and the correct balance between the membrane permeability and selectivity could be used to improve their inhibition potential against Mycobacterium tuberculosis [47]. Delaine et al., in 2010 reported that shortened adducts of isoniazid-NAD containing a lipophilic fragment could act as effective bi-substrate inhibitors of InhA and antimycobacterialcompounds[48]. Such truncated adducts resembling the active metabolite of isoniazid, were designed and synthesized combining a lipophilic hydrocarbon resembling theENR substrate and thenicotinamide group of NAD. Both classical nucleophilic substitutions and Suzuki-Miyaura cross-coupling reaction were used to introduce the lipophilic fragment. The synthesized compounds were able to show significant inhibition of InhAalong with promising antimycobacterial activities. Three series based on pyridinium, pyridine, and 1,4-dihydropyridines (Fig. 3) were synthesized that were simplified derivatives of ENR-NAD adduct structurally. More active compounds were formed on introduction of a lipophillicfragment into nitcotinamidehemiamidal framework. Compounds (25), (26), (27), (28) and (29) showed a significant InhA inhibitory activity. Several in silicoQSAR studies were carried out to generate pharmacophore models using different scaffolds for Inh A inhibitors. Furthermore molecular docking analysis was done toexplore the interactions of 28 arylamide derivatives with the target enzyme [49]. CoMFA, CoMSIA, and HQSAR studies were employed to determine the structure activity relationship of these arylamide derivatives. It was found out that the critical structural features for the the searylamides to fit into the binding pocket of InhA included H-bond formation between the oxygen of the amide carbonyl and the hydroxyl substituents of Tyr158 and nicotinamide ribose; hydrogen bond-formation between oxygen of pyrophosphate and NADH with ring A meta and orthohydrogens; appropriate size of ring A and ring B substituents for an optimum distance for hydrogen bond interactions with NADH and Tyr158; hydrophobic ring B interactions with Met232, Ala157, Ile215,Met199, Leu218, Pro193,Val203, Trp222 and Ile202. A pharmacophore model was built taking a conformation of indole-5-amide inhibitor (Genz 10850) bound to InhA using Ligand Scout (PDB code: IP44) [50]. This model was then applied to forty structurally varied arylcarboxamideanalogues synthesized by Kuo et al [45] to align them in order to identify the bioactive conformation. Based on pharmacophore alignment (CoMFA) and CoMSIA analysis of arylcarboxamides-based inhibitors of InhAand graphical interpretation of the data the information about the critical structural attributes of thesearylcarboxamidesfor inhibition of InhA were obtained. The developed pharmacophore model could thus be applied to discover new hits by virtually screening commercial databases and then calculate the activities. Deraeve C. et al.,[51] designed and synthesizedazaisoindolinonebased compounds containing a lipophilic chain as inhibitors of InhA and as anti-tubercular agents. SAR studies were done to establish the appropriate size and position of the lipophilic chain around the azaisoindolinonemoiety. It was found that the bioisosteric substitution of ether link, the suppression of the phenyl group, and alkylation of the thehemiamidal nitrogen and tertiary hydroxyl affected the inhibition activity. This insightful information could thus be employed for further modification of the azaisoindolinone framework for development of new anti-TB agents.
 |
Figure 3 Click here to View figure |
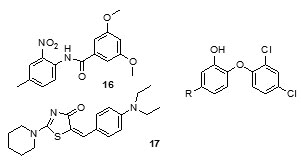
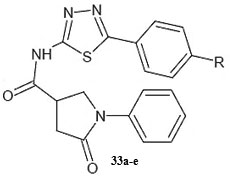
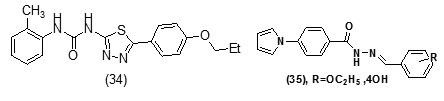
It was observed that aryl azaisoindolinone framework (fig.4) has a tendency to form epimers at the C-7 carbon by openingof ring. SAR analysis around the parent azaisoindolinone scaffold (30) was performed to help in confirming some pharmacophore elements. It was observed that presence of phenyl ring with lipophilic chainon theazaisoindolinonemoiety resulted in the most effective molecular interactions with ENR. Furthermore the presence of sulfur or oxygen atom bound long carbonchain (12 or 18)on the phenyl ring at meta position and a C-7positioned free tertiary hydroxyl resulted in enhanced interaction with InhA. These structural modifications led to an enhanced inhibition of M. tuberculosisgrowth and also in improving the activity against InhA.Compounds (31) and (32) were found to exhibit better inhibition ofInhA and superior activity against M. tuberculosis growththan the parent molecule (30). On the basis of docking studies and oral bioavailability scores using Lipinski’s rule evaluation, a series of 5-oxo-1-phenyl-N-(5-substituted phenyl-1,3,4-thiadiazol-2-yl)pyrrolidine-3-carboxamides (33a-e) were designed and synthesized as mycobacterium tuberculosis ENR (InhA) inhibitors. Theydispalyed good anti tubercular activity confirming them to be promising candidates as MtbInhAinhibitors[52]. QSAR study was carried out on a twenty member series of 1,4- dihydropyridine derivatives reported as inhA inhibitors to provide reliable clues for further optimization of 1,4-dihydropyridine pharmacophore as effective antitubercular agents by Deshmane S et al.,[53]. An anti-tubercular screening model of pyrrole derivatives was constructed by Zhou, S et al.,[54] by molecular docking with InhA. An ensemble-docking processemploying the AutodockVina software was carried out by Stigliani JL et al.,[55]with four X-ray crystallographic structures.ofInhA. Five inhibitors ofInhAwere docked sequentially in the cavity of the proteinfor the substrate. Comparison of the calculated conformations to the crystallographic structures was employed to assess and validate the efficiency of the docking. The bioactive conformation of each ligand was retrieved using interaction energies combined with the multiple receptor conformations approach. QSAR studywas performed by Jain A K et al.,[56]using a data obtained for a series of 23 compounds as inhibitors of InhA (Endeet al., 2008) [57] in order to facilitate the design ofdiaryl ether analogues as ENRinhibitors by making modifications in the B-ring. The information obtained from a study carried out by Kumar V et al.,[57] that direct inhAinhibitors upon binding cause conformational changes in the binding loop for the substrate (SBL) of InhA, could be utilized as a parameter for enhanced binding interaction with InhA. Based on 36 available Mt InhA crystal structures a model of a 3-D pharmacophore was built by Pauli I et al.[58]. So as to identify novel InhA inhibitors from the ZINC database by selecting a library of molecules, the authors employed two different approaches of in silicoligand-screening. 3-D pharmacophoremodel was first developed using 36 available MycobacteriumInhA crystal structures. Four pharmacophoric points were employed by combining ligand-based and structure-based data in order to get molecules which could compliment the binding features of MycobacteriumInhA substrate-binding cavity. To study the binding mode and affinity of the chosen molecules another approach involving theuseoffour well known docking programs which employ different search algorithms was made.After analyzing the results, out of the six ligands selected for in vitro analysis three molecules were found to exhibit inhibition with IC50 values in the range of 24 (±2) μM to 83 (±5) μM. The most potent compound (34) displayed an uncompetitive inhibition mode to 2-trans-dodecenoyl-CoA substrates and NADH and, with Ki values of 20 (±2) μM and 24 (±3) μM respectively. A study conducted by Kumar U C et al.,[59] led to generation of a quantitative pharmacophore model by making use of known ENR inhibitors, which was further authenticated by employing a large test set of compounds. 400,000 compounds from a database were screened using the validated pharmacophore model and 25,000 hits were retrieved. Rapid overlay of structures (Open Eyertm) with potent ENR inhibitors was employed to grade these hits based on shape and other feature similarity, followed by subjecting them to docking. This led to selection of thirty compounds with greater than eight chemotypes. The compounds were procured commercially and evalvatedfor inhibition of InhA. Twenty eight out of the selected thirty two compounds at a concentration of 10 mM were found to demonstrate 10-38% inhibition against InhA. Srinivaset al., reported synthesis of a series of 4-(4-pyrrol-1-yl/2,5-dimethyl-4-pyrrol-1-yl) benzoic acid hydrazidederivatives, derived oxadiazoles and azines. Docking of these molecules into InhA revealed binding conformation of these molecules and the vital interactions with the target. It was observed that the compounds (35), (36), (37), (38), (39) and (40) interacted with InhA enzyme more efficiently [60]
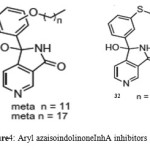 |
Figure 4 Click here to View figure |
Design and synthesis of a series of twenty eight 2-(4-oxoquinazolin-3(4H)-yl)acetamideanaloguesas significant MTBInhA inhibitors has been reported [61].Compound (41) (R1=Cl, R2= Phenyl R3=CH3 ) was found to be the most potent derivativeshowing88.12% inhibition of ENR at 10 mMand an IC50 of 3.12 mM. Furthermore, compound (41) was also found to inhibit drug sensitive M. tuberculosisexhibiting an MIC of 4.76 mM and did not show any cytotoxicity at 100 m M. Based on earlier reported lead (42)against the MTBInhA a series of twenty seven substituted 2-(2-oxobenzo[d]oxazol-3(2H)-yl)acetamideanalogues were synthesized. 2-(6-nitro-2-oxobenzo[d]oxazol-3(2H)-yl)-N-(5-nitrothiazol-2-yl)acetamide (43) (R1= Nitro, R2=5-Nitrothiazol-2-yl) exhibited the most potent inhibiton showing an IC50 of 5.12 ± 0.44 µM against M. tuberculosis ENR. It was reported to inhibit the drug sensitive M. tuberculosiswith an MIC of 17.11 µM and did not show any-cytotoxicity at 100 µM[62]. 3D-QSAR analysis and a molecular dynamics simulations study was used to elucidate the underlying structural requirements of diphenyl ether derivatives for inhibition of InhA. 3D-QSAR CoMSIA models were also employed to identify the structural basis for the inhibition based on available experimental binding data of di-Ph ethers. For the calculation of binding free energies four representative compounds were selected and employed for MD simulations with InhA. Obtained results showed that the presence of R1 bulky groups on the Ph A ring of the molecules would fit well in the hydrophobic pocket formed by residues Met155, Phe 149, Ala157, Pro156, Tyr158, Pro193, Val203,Met199, Ile215,Leu207 and Leu218. Furthermore the presence of small hydrophilic substituents at the R3 and R4 positions of thePh B rings would result in hydrogen bonds interactions with Gly96 and Met98 residues, resp. At R2 position the presence of small groups with simultaneous hydrophobic or hydrophilic properties are required to form a favorable interaction with the Me side chain of Ala198 and the pyrophosphate moiety of NAD+, resp. This data helped in the further design of novel and more potent di-Ph ether-based inhibitors against mtInhA[63, 64].
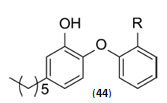
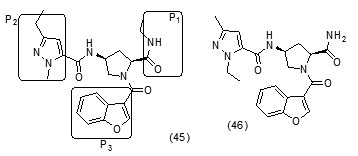
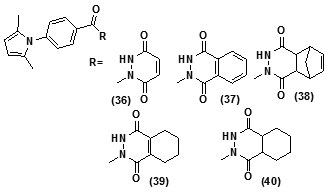
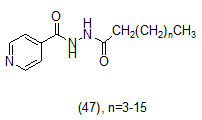
The study carried out by Pan Panet al., [65, 66] demonstrated that B-ring modified diaryl ethers (44 a) (R=CH3) and (44 b) (R=Cl)} showed time-dependent InhAinhibition and possessed antibacterial activity when administered intraperitoneally, against TB infection in a murine model. The study proposed that residence time of these compounds on the enzyme was related to their efficacy. Based on the diaryl ether scaffold twenty compounds were synthesized, with anA-ring which was hexyl substituted and with several alterations in the B-ring. Following which the synthesized compounds were screened for InhA enzyme inhibition and antibacterial activity using acell based platform. Furthermore crystallographic analysis demonstrated that the slow-binding inhibitors (44c) (R=Br) and (44d) (R=CN) resulted in a more controlled and closed ENR substrate binding loop. It was also observed that on the B-ringthebulk of orthosubstituents highly influenced the activity of the diaryl ether derivatives. The activity was significantly reduced if greater than two heavy atoms where present in the group. For this group the presence of non-hydrogen bonding donor groups conferred better activity within the suitable size limits. On insertion of substituents on both orthopositions aloss of activity was observed due to a conformation which according to the docking studies, resulted inuncomplimentary van der Waals interactions with InhA. Compound (45)havinggood inhibition potential but low antitubercularactivity was developed by using encoded library technology (ELT) for the discovery of direct InhA inhibitors [67].With an objective of improving the potency, the proline core wasused as template and was systematically investigated by making sequential modification son the three possible positions to generate chemical diversity. This approach was quickly able to afford structure activity relationship information. Subsequent to optimizing combinations of the most suitable structural features at each position, P3 turned out to be the least complaint position for structural activity relationship manipulations. It was found that for potent enzymatic activity,the substituent group at position P3played a key role. Diethylpyrazoletranspired as the most suitable substituent at position P2 for potent anti-tubercular activity. Ability to improve physicochemical and ADME properties was provided by the SAR analysis around P1. The optimized lead compound (46) was synthesized by the combination of best structural features. N-acylatedisonicotinic acid hydrazideanalogues (47) were synthesized [68] as potential InhAinhibitors and were found to inhibit the growth of Mycobacterium tuberculosis H37Rv and two human clinical isolates of mycobacterium. As compared with isoniazid the synthesized hydrazide derivatives exhibited fair to excellent inhibition against the tested strains and were found to be non-toxic with LD50 values ranging from 3898 to 5000 mg/kg body wt.Pyrrolyloxadiazole derivatives[69]were synthesized followed by their molecular modeling and antitubercular evaluation.
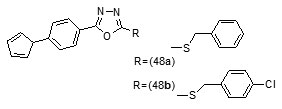
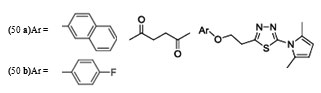
The result indicated that, the synthesized compounds exhibited moderate antitubercular activity. In docking analysis it was observed that compounds (48a) and (48b) bound tightly to the Enoyl ACP-reductase enzyme. Fifty two, novel pyrrole hydrazine derivatives [70]were synthesized and screened against the target enoyl-ACP reductase. Surflex-docking method was used to explorethe binding mode of the synthesized compounds at the active site of InhA. The binding model suggested formation of one or two hydrogen bonds between the InhAand pyrrolehydrazones. The most potent compound (49) (MIC 0.2 mg/mL) was found to exhibit H- bonding interactions similar to that of Triclosan, i.e., with Tyr158 and NADP. Sixty eight novel pyrrolyl substituted aryloxy-1,3,4-thiadiazoles have been synthesized by three-step optimization processes [71]. For pyrrolyl substituted aryloxy-1,3,4-thiadiazole series of InhA inhibitors 3D-QSAR was established using the comparative molecular field analysis (CoMFA) and docking studies were also carried out using the crystal structure of InhA. 3D contour plots on analysis helped to explore the effect of different substituents at different positions of the common scaffold. Results obtained through in silico methods were substantiated in invitro testing of ligands using biological assays. Among the compounds investigated, (50a) and (50b) displayed significant activities (3.125 and 6.25µg/mL) against M. tuberculosis H37Rv strain. Docking simulation studies revealed that these thiadiazoles were mainly bound to the substrate binding site of enoylreductase and the scoring function for all the derivatives was similar or higher than that of the standard inhibitor. Designed structures have shown interactions with the substrate binding site of InhA, confirming their high inhibitory potency, depending on the type of aryl ring modification.Drug repositioning approach to drug discovery and development was applied in finding potential inhibitors of enoylreductase[72].
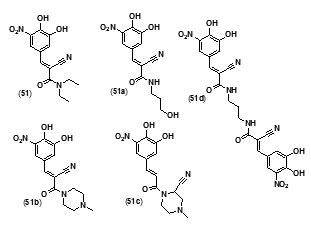
Entacapone (51), a drug for Parkinson’s disease, was also found to inhibit InhA enzyme. A compd. database was scoured to search for entacapone-like structures, which were then filtered based on LibDock scores. The hits were subsequently docked into InhA binding site by the use of C Docker protocol and their binding energies were calculated. The results showed that the dimer, and an alcoholic and piperazine derivatives of entacapone are potential inhibitors of InhA. H bonding and p-p interactions with nicotinamide adenine dinucleotide (NAD) at the binding pocket were found to be the salient features in binding interactions. The four entacapone analogs (51a-d) exhibited greater binding affinity with InhA compared to entacapone itself (fig. 5).
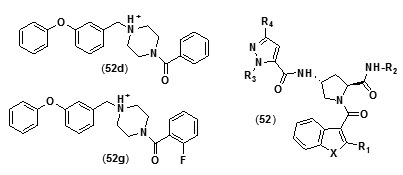
Benzofuranpyrrolidinpyrazole derivatives (N-((3R,5S)-1-(benzofuran-3-carbonyl)-5-carbamoylpyrrolidin-3-yl)-1H-pyrazole-5-carboxamide derivatives (52) were reported as direct and strong inhibitors of InhA. With the purpose of investigating the basis for the potency of these compounds against InhA and tuberculosis with respect totheir structure, comparative molecular similarity indices analysis (CoMSIA) and comparative molecular field analysis (CoMFA) was carried out using IC50 and MIC90 values of a set of thirty four pyrazole,pyrrolidine and benzofuran derivatives. In order to attain favorable IC50 values the core structural motif of the compound used as template was found to be crucial, whereas the R2 substituent was found to play a key role in enhancing MIC90 values without negative effects on IC50values[73]. A novel series of diphenyl ether analogues was synthesized on the basis of matched molecular pair (MMP) method [74], following which in silico screening was employed to calculate the binding energies of these analogues. Ten unique candidate compounds were identified.They were then evaluated formtInhA inhibition followed by assessment of their in vitro antibiotic activity against mycobacteria.
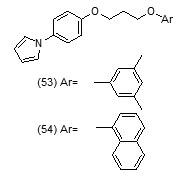
Their toxicity level on both mammalian cellsandintestinal bacteria was also determined. Compound (52d) with phenyl group substitution and compound (52g) with 2-fluorobenzyl group as substituents were found to show better inhibition of mtInhAin comparison to the compounds which have a furyl group as terminal substituent.Compound (52g) was found to show a relatively lower inhibition of the growth of the mycobacteria. In an effort to develop more potent Inh A inhibitors a series of pyrrolylphenoxy analogues bearing alkoxygroup as linker were prepared and assessed against Mycobacterium tuberculosisfor anti-tubercular property[75].
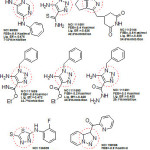 |
Figure 6 Click here to View figure |
The optimized pharmacophore model was developed and validated. Compound (53) showed hydrogen bond interactions with Thr196, Met199, Tyr158, and NAD+ that fitted closely into the binding site of InhA whereas compound (54) was found to show the H-bond with NAD+. Acceptor groups with benzene ring and the alkoxy linker bridge were found advantageous for anti-tubercular activity. Using phenotypic high-throughput whole-cell screening 4-hydroxy-2-pyridones have been reported to constitute a new class of direct InhA inhibitors [76]. These compounds were found to show potent bactericidal activity against common isoniazid-resistant TB clinical isolates. It was revealed in biophysical studies that these compounds bound specifically to InhA in an NADH (reduced form of NAD)-dependent manner and blocked the InhA binding pocket. The lead compound NITD-916 (55) directly inhibited InhA dose-dependently. It was also found to show efficacy in in vivo acute and established mouse models of Mycobacterium tuberculosis infection. Virtual screening of compounds from GO FAM project of NCI was performed against InhA in order to discover new scaffolds for further development The structural features namely, requirement of compounds to have an anticipated free binding energy ≤−8.0 kcal/mol; to form a minimum of two H bonds with the active site and to base-stack with NAD cofactor were used as interaction-based filters. Eight of the 16 soluble compounds did not involveinitial activation by KatG and exhibited modest inhibition of (27−71% at 100 μM) [77].
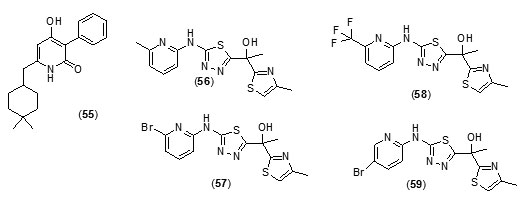
The two best inhibitors that were discovered showed Ki values of 54 and 59 mM, respectively and were fragment-sized analogues (fig.6).Novel inhibitors discovered in this study had very little similarity to known inhibitors of InhAstructurally and thus has led to expansion in the diversity of chemical spacefor future exploration. Tetracyclic thiadiazole[78]based direct and competitive inhibitors ofInhAwere taken up for further exploration as they were found to have anti-tubercular activity in vivoin murine models. On the basis of this template shortened analogues with three aromatic rings were explored. The analogues (56), (57), (58), (59) and (60) were found to not only exhibit nanomolarInhA potency and anti-mycobacterial activity with submicromolar potency but they also exhibited better physicochemical propertiesas compared with the tetracyclic thiadiazoles.
Compound, (57) was reported as the most potent molecule, while as the pure S-enatiomer of (57) showed significant in vivo efficacy.
Conclusion
The data presented here, firmly reiterates that the path way for fatty acids biosynthesis is an important target for discovery of drugs for tuberculosis. Given the continuing and developing problems of drug resistance, and in particular multid rug-resistance (MDR), aggressive efforts are required for development of newanti-tubercular drugs in order to combat the bacteria that are resistant to current therapeutics. Several approaches such as de novo design, computational simulation and side activity optimization selectively have been employed for development of inhibitors of FAbI showing good potency, but despite that very few molecules have attained clinical development. More efforts need to be concentrated in enhancing thein vivo properties of the molecules,especially the properties which affect ADME, andother important parameters which influence their potency for development of more efficient InhA inhibitors.
References
- Babu,Giridhara R.; Laxminarayan, R. Tuberculosis 2012, 92 (4), 301-306.
- World Health Organization. WHO progress report20112011.
- Kaur, R.; Kaur, H. Oriental Journal of Chemistry 2015, 31 (1), 597-600.
- Billones, J. B.; Carrillo, M.C.O.; Voltaire G. O.;Stephani, J. Y. Macalino, I. A. E.;Bernadette A. S. J.; Oriental Journal of Chemistry 2014, 29(4), 1457-1468..
- Takayama, K.;Wang,L.; Hugo, L. D.;Antimicrobial agents and chemotherapy 1972, 2 (1), 29-35.
- Banerjee, A.; Eugenie, D.;Annaik, Q.;Balasubramanian, V.; Kyung, S.U.Science 1994, 263 (5144), 227-230.
- Annaiek, Q.; Sacchettini, J.C.;Dessen,A.;Vilcheze, C.;Bittman, R.; Jacobs, W. R. ;Blanchard,J. S;Biochemistry 1995, 34 (26), 8235-8241.
- Sheldon, M.;Bai, G.H.;Suffys, P.; Gomez,L. P.;Fairchok,M.;Rouse, D.;Journal of Infectious Diseases 1995, 171 (4), 954-960.
- Musser, J. M.;Kapur, V., Williams, D. L.; Kreiswirth, B.N.;Soolingen, D. V.; Embden, .D.V.;Journal of infectious Diseases 1996, 173 (1), 196-202.
- Zhang, Y.;Heym, B.; Allen, B.; Young, D.; Cole, S. Nature1992, 358(6387), 591-593.
- Davies, A. P.;Billington,O. J.; Bannister, B. A.; Weir, W. R. C.; McHugh, T. D.; Gillespie, S. H.;Journal of Infection 2000, 41 (2), 184-187.
- Khisimuzi,M.;Slayden, R.A.; Zhu, Y.Q.; Ramaswamy, S.; Pan, X.; Mead, D.; Crane, D.D; Musser, J. M.; Barry, E. A. Science 1998, 280 (5369), 1607-1610.
- Slayden, R.A.; Richard, E. L.; Barry, C. E. Molecular microbiology 2000, 38 (3), 514-525.
- Larsen, M. H.;Vilchèze, C.; Kremer, L.;Besra, G. S.; Parsons, L.;Salfinger, M.; Heifets, L; Hazbon, M. H.; Alland, D.; Sacchettini, J.C.; Jacobs, W. R .Jr. Molecular microbiology 2002, 46 (2), 453-466.
- Kremer,L.; Dover, L.G.; Morbidoni, H.R.;Vilchèze, C.;Maughan, W. N.;Baulard, A.; Tu, S.C.;Honore, N.; Derectic V., Sacchettini, J .C.; Locht, C.; Jacobs, W. R .Jr. ; Besra G.S. Journal of Biological Chemistry 2003, 278 (23), 20547-20554.
- Andrea, D.;Quemard, A.; Blanchard, J. S.; Jacobs, W. R .Jr. ; Sacchettini, J .C. Science 1995, 267 (5204), 1638-1641.
- Kai, J.; King,D.S.;Schultz, P.G.Journal of the American Chemical Society 1995, 117 (17), 5009-5010.
- Marcinkeviciene, J. A.; Magliozzo, R. S.; Blanchard, J. S.Journal of Biological Chemistry1995, 270 (38), 22290-22295.
- Basso, L. A.;,Zheng,R.;Blanchard, .S. Journal of the American Chemical Society1996, 118(45), 11301-11302.
- Annaik, Q.;Dessen, A.;Sugantino, M.; Jacobs, W. R.;Sacchettini, J. C.; Blanchard, J. S. Journal of the American Chemical Society 1996, 118 (6),1561-1562.
- Rozwarski, D. A.; Grant, G.A.; Barton, D. H. R.; Jacobs, W. R.;Sacchettini, J. C. Science 1998279 (5347), 98-102.
- Richa, R.; Whitty, A.; Tonge, P.J. Proceedings of the National Academy of Sciences 2003, 100 (24), 13881-13886.
- Hao, L.;Tonge, P. Accounts of chemical research 2008, 41 (1), 11-20.
- Stoeckle, M. Y.; Guan, L.;Riegler, N.; Weitzman, I.; Kreiswirth, B.; Kornblum, J.;Laraque, F.; Riley, L. W. “ Journal of Infectious Diseases 1993, 168 (4) , 1063-1065.
- Ramaswamy, S.V.; Robert Reich, R.; Dou, S. J.;Jasperse, L.; Pan, X.; Wanger, A.; Quitugua, T.; Graviss, E.A. Antimicrobial agents and chemotherapy 2003, 47 (4), 1241-1250.
- Hao, L.; England, K.;Ende, C.A.;Truglio, J. J.;Luckner, S.; Reddy, B.G.; Marlenee, N.L.; Knudsen, S. E.;Knudsen, D. L.; Bowen, R.A.; Kisker, C.; Slayden, R.A.; Tonge, P. ACS chemical biology 2009, 4 (3), 221-231.
- Lu, X.Y.; Chen, Y. D.; Jiang,Y.J.;You, Q. D. European journal of medicinal chemistry 2009, 44 (9), 3718-3730.
- Ashutosh, K.;Siddiqi, M. A. Journal of molecular modeling2008, 14(10), 923-935.
- Kinnings, S. L.; Liu, N.;Buchmeier, N.; Tonge, P.J.; Xie, L.; Bourne, P.E. PLoSComputBiol 2009, 5(7), 1000423.
- Rao, S.; Vijayakrishnan, G.;R.;Kumar.M.Chemical biology & drug design 2008, 72 (5), 444-449.
- Sullivan, T. J.;Truglio, J. J.; Boyne, M.E.; Novichenok, P.; Zhang, X.; Stratton, C. F.; Li, H. J.; Kaur, T.; Amin, A.;Johnson, F.; Slayden, R. A.; Kisker, C.; Tonge. P. ACS chemical biology 2006, 1 (1), 43-53.
- Kuo, M. R.; Morbidoni, H.R.; Allandll, D.; Sneddon, S. F.; Gourlie, B, B.; Staveski, M. M.; Leonard, M.; Gregory, J. S.; Janjigian, A.D.; Yee, C.; Musser, .M.; Krieswirth, B,; Iwamoto, H.; Perrozo, R.; Jacobs, W.R.; Sacchettini, J.C.; Fidock, D.A. Journal of Biological Chemistry 2003, 278 (23), 20851-20859.
- Hao, L.;Tonge, P.J.;Current opinion in chemical biology 2010, 14 (4), 467-474.
- Copeland, R. A., Pompliano, D. L.;Meek, T.M. Nature reviewsDrug discovery2006, 5 (9), 730-739.
- Tummino, P. J.; Copeland, R.A.; Biochemistry 2008, 47 (20), 5481-5492.
- Swinney, D. C. Nature reviews Drug discovery 2004, 3 (9), 801-808.
- Andrew, J.;Sneddon, S. F.; Staveski, M. M., Yee, C.; US6372752 B1 2001, 56.
- Sylvain, B.;Génisson, V.B.; Gornitzka, H.; Bernadou,.;Meunier, B.;Organic&biomolecular chemistry 2005, 3 (4), 666-669.
- He, X.;Akram, A.; Stroud, R.; Montellano, P.R.D.O. Journal of medicinal chemistry 2006, 49(21), 6308-6323.
- He, X.;Akram, A.; Paul,R.Bioorganic& medicinal chemistry 2007, 15 (21), 6649-6658.
- Tonge, P.J.; Sullivan,T.; Johnson, F.;Research Foundation of State University of NY, USA2006, 10.
- Boyne, M. E.; Sullivan, T..; Lu, H.;Gruppo, V.;Heaslip, D.; Amin, A. G.;Chatterjee, D.; Lenaerts, A.;Tonge, P.J.; Slayden, R.A.. Antimicrobial agents and chemotherapy2007, 51 (10), 3562-3567.
- Wall, M.D.; Oshin,M.; Chung, G.A.C.; Parkhouse, T.; Gore, A.; Herreros, E.; Cox, B.; Duncan K.; Evans, B.; Everett, M.; Mendoza, M. Bioorganic & medicinal chemistry letters 2007, 17 (10), 2740-2744.
- Ende, C.W. A.; Knudson, S. E.; Liu, N.; Childs,J.; Sullivan,T.J.; Boyne,M.; Xu, H.; Gegina, Y.; Knudsen, D.L.; Johnson, F.; Peloquin, C.A.; Slayden, R.A.; Tonge, P.J. Bioorganic & medicinal chemistry letters 2008, 18 (10), 3029-3033.
- Freundlich, J. S.; Wang, F.; Vilchèze, C.;Gulten, G.; Langley, R.;Schiehser, G.A.; Jacobus, D. P.; Jacobs, W. R.; Sacchettini..C.;ChemMedChem 2009, 4 (2), 241-248.
- Luckner, S. R.; Liu, N.; Ende, C.A;Tonge, P.J.;Kisker, C. Journal of Biological Chemistry 2010, 285 (19), 14330-14337.
- Punkvang, A.; Saparpakorn, P.;Hannongbua, S.; Wolschann, P.; Beyer,A;PornpanPungpo, P.; European journal of medicinal chemistry 2010, 45 (12), 5585-5593.
- Delaine, Tamara, VaniaBernardes-Génisson, AnnaïkQuémard, Patricia Constant, Bernard Meunier, and Jean Bernadou. European journal of medicinal chemistry2010, 45 (10), 4554-4561.
- Auradee, P.;Saparpakorn, P.;Hannongbua, S.;Wolschann, P.; Berner, H.;Pungpo, P.;MonatsheftefürChemie-Chemical Monthly 2010, 141 (9), 1029-1041.
- Lu, X.Y.; Chen,Y.D.;You, Q.D.;Chemical biology & drug design 2010,75 (2), 195-203.
- George, S. O. N. I. A.;Kochupappy, R.T.International Journal of Pharmacy and Pharmaceutical Sciences 2013, 3 (4), 280-284.
- Sonali, D.; Asgaonkar, K. D.; Patil, S.; Landge, D.; Chitre, T.S. Inventi Rapid: Med Chem 2012, 208
- Shanshan, Z.X., Zhenmin,M.Computers and Applied Chemistry2012, 29(4), 427-430.
- Stigliani, J.L.; Génisson, V.B.;Bernadou, J.; Pratviel, G.Organic&biomolecular chemistry 2012, 10 (31), 6341-6349.
- Abhishek, J.K..; Veerasamy, R.; Vaidya, A.;Kashaw, S.;.MouryaV. K.; Agrawal, R. K. Medicinal Chemistry Research 2012, 21 (2), 145-151.
- Kumar, V.;Sobhia, M.E.;International journal of computational biology and drug design 2013, 6 (4), 318-342.
- Ivani, P.; Santos, R.N.C.; Rostirolla, D.C.; Martinelli, L.K.; Ducati, R. G.; Timmers, L. F.S.M.; Basso, L.A.; Santos, D.S.; Guido, R.V.C.; Andricopula, A.D.; Souza, O.N.D. Journal of chemical information and modeling 2013, 53 (9), 2390-2401.
- Kumar, U. C., Kumar S.B.V.S.; Mahmood, S.;Sahu, P.K.; Sridevi, J.P.; Ballell, L.; Gomez, D.A.; Malik, S.;Sarma, J.A.R.P. Future medicinal chemistry 2013, 5 (3), 249-259.
- Shrinivas, J. D.;Uttam A.M.; Pansuriya, K.; Aminabhavi, T. M.; Gadad, A.K.;Journal of Saudi Chemical Society 2013, XXX, XXX-XXX
- Pedgaonkar, G.S.;Sridevi, J.P.,Jeankumar, J. U.; Saxena, S.; Devi, P.B.; Renuka, J. ; Yogeeswari, P.;Sriram, D.European journal of medicinal chemistry 2014, 86, 613-627.
- Pedgaonkar, G.S.; Sridevi, J.P.,Jeankumar, J. U.; Saxena, S.; Devi, P.B.; Renuka, J. ; Yogeeswari, P.;Sriram, D. Bioorganic & medicinal chemistry 2014, 22 (21), 6134-6145.
- Kamsari, P.; Koohatammakun, N.; Srisupan, A.; Meewong, P.; Punkvang, A.; Saparpakorn, P.;Hannongbua, S.; Wolschann. P.; Prueksaaroon S, Leartsakulpanich U, Pungpo, P.; SAR and QSAR in Environmental Research 2014, 25(6), 473-488.
- Kamsari, P.; Punkvang, A.; Hannongbua, S.; Saparpakorn, P.; Pungpo, P.;RSC Advances 2015, 5 (65), 52926-52937.
- Pan P, Knudson, S. E.;Bommineni, G.R.; Li, H.J., Lai, C.T.; Liu, N.; Diaz, M.G.; Simmerling, C.; Patil, S.S.; Slayden, R. A.; Tonge, P.J. ChemMedChem 2014, 9 (4), 776-791.
- Pan, P.; Liu, N.; Lai, C.T.; Shah, S.;Bommineni, G. R.; Knudson, S.; Slayden, R. A.; Tonge, P.J. In Abstracts Of Papers Of The American Chemical Society, 2011,242, 1155.
- Lourdes, E.; Keefe, H. O.;Neu, M.;Remuiñán, M. J.; Patel, A. M.;Guardia,A.; Davie, P. D.;Macais, N. P.; Yang, H.; Convey, M. A.; Messer, J.A.; Herran, E.P.; Centrlla, P. A.; Gomez, D.A.; Clark, M.A.; Huss, S.; Donovan, G, K. O.; Muro, F.O.; McDowell, W.; Cataneda, P.; Muendel, C.A.;Pajk, S.; Rullas, J.; Barturen, I.A.; Ruiz, E.A,;Losana, A.M.;Ballell, L, Pichel, J.C.; Evindar, G. J. Med. Chem 2014, 57 (4), 1276-1288.
- Kumar, H. S. N.; Parumasivam, T.; Ibrahim, P.;Asmawi, M. Z.;Sadikun, A.; Medicinal Chemistry Research 2014, 23 (3), 1267-1277.
- Shrinivas, J. D.;Sheshagiri, R. D.; More.U.A.; Rai, S.; Kulkarni, V. H.; Indo American Journal of Pharm Research 2014, 12 (4). 2323-2338
- More, U. A.; Joshi, S. D.; Aminabhavi, T. M.;Gadad, A. K.; Nadagouda, M. N.; Kulkarni, V.H.; European journal of medicinal chemistry 2014, 71, 199-218.
- Joshi, S. D.; More, U.A.; Koli, D.; Kulkarni, M. S.; Nadagouda, M. N.; Aminabhavi, T.M.;Bioorganic chemistry 2015, 59, 151-167.
- Cordova, D.L.M.; Abeul, R.J.D.; Galingana, M.O.;Villanueva, L.A.;Billones, J.B.J. Chem. Pharm. Res., 2015, 7(5), 636-642.
- Pharit, K.; Punkvang, A.; Hannongbua, S.; Saparpakorn, P.; Pungpo. P. RSC Advances 2015, 5(65), 52926-52937.
- Hironori, K.; Koseki, Y.; Taira, J.; Umei, T.; Komatsu, H.; Sakamoto, H.; Gulten, G.; Sacchettini, J. C.; Kitamura, M.; Aoki, S.European journal of medicinal chemistry 2015,94, 378-385.
- More, U, A.; Joshi, S. D.;Aminabhavi, T. J; Kulkarni, V. H.;Badiger, A.M.; Lherbet, C.European journal of medicinal chemistry 2015, 94, 317-339.
- Manjunatha, U. H.; Rao, S. P. S.;Kondreddi, R.R.; Noble, C.G.; Camacho, L.R.; Tan, B.H.; Ng, S.H.; Ng, P.S.; Ma, N. L.; Lakshminarayana, S.B.;Herve, M.; Barnes, S.W.; Yu, W.; Kuhem K.; Blasco, F.; Beer, D.; Walker, J .R.; Tonge, P.J.; Glynne, R.; Smith, P.W.; Diagana. T. T. Science translational medicine 2015, 7 (269): 269ra3-269ra3.
- Perryman, A. L.; Yu, W.; Wang, X.; Ekins, S.; Forli, S.; Li, S. G.; Joel S. Freundlich, J. S.;Tonge, P. J.; Olson, A. J. Journal of chemical information and modeling 201555 (3), 645-659.
- Roman, S.; Sosic, I.; Živec, M.; Menendez, R. F.; Turk, S.; Pajk, S.; Gomez, D.A.; Triconado, J. R.; Barturen, I. A.;Barros, D.; Pages, L.B.; Young, R..; Encinas, L.; Gobec, S. Journal of medicinal chemistry 2014, 58 (2), 613-624.

This work is licensed under a Creative Commons Attribution 4.0 International License.









