Synthesis, Characterization, and Antibacterial Activity of Cu(II), Ni(II), and Zn(II)Complexes of 14-Membered MacrocyclicTetraazaLigand
Siti Fairus M. Yusoff,1*Ameera Aqeela Salleh Huddin1,Latifah M. Yusoff1, Bohari M. Yamin1, NazlinaIbrahim2,and Ong Wei Leng2
1School of Chemical Sciences and Food Technology,
2School of Biosciences and Biotechnology,
Faculty of Science and Technology, UniversitiKebangsaan Malaysia, 43600 Bangi, Selangor, Malaysia.
Corresponding Author Email: sitifairus@ukm.edu.my
DOI : http://dx.doi.org/10.13005/ojc/310356
Article Received on :
Article Accepted on :
Article Published : 15 Sep 2015
The macrocyclic5,5,7,12,12,14-hexamethyl-1,4,8,11-tetraazacyclotetradeca-7,14-dienium bromide salt (LBr) was synthesised for use in complexation reactions with various metal salts of copper, nickel, and zinc. Twelve complexes were synthesised; five of them were successfully obtained in a crystal form that was suitable for X-ray crystallographic analysis. All the complexes were analysedby microelemental analysis, Fourier Transform infrared spectroscopy, ultraviolet-visible spectroscopy,and magnetic susceptibility. The metals formed coordination bonds to the four aza nitrogen atoms,during the process of which the azomethine nitrogen atoms were deprotonated. The cationic charge in each complex was balanced with a bromide anion from the macrocyclic ligand. Deprotonation of the azomethine nitrogen atoms led to the formation of an acid/base complex with the metal salts.Preliminary studies for biological activity, based on the minimum inhibitory concentration (MIC) method, indicated that the macrocyclic complexes showed greater antibacterial activity than the bromide macrocyclicligand, which showed lower MIC values with S. aureus(6.3 mg/mL) and E. aerogenes and B. Subtilis (3.1 mg/mL).
KEYWORDS:Tetraaza; Macrocyclic; Copper; Nickel; Zinc Complexes; Antibacterial
Download this article as:| Copy the following to cite this article: Yusoff S. F. M, Huddin A. A. S, Yusoff L. M, Yamin B. M, NazlinaIbrahim, Leng O. W. Synthesis, Characterization, and Antibacterial Activity of Cu(II), Ni(II), and Zn(II)Complexes of 14-Membered MacrocyclicTetraazaLigand. Orient J Chem 2015;31(3). |
| Copy the following to cite this URL: Yusoff S. F. M, Huddin A. A. S, Yusoff L. M, Yamin B. M, NazlinaIbrahim, Leng O. W. Synthesis, Characterization, and Antibacterial Activity of Cu(II), Ni(II), and Zn(II)Complexes of 14-Membered MacrocyclicTetraazaLigand. Orient J Chem 2015;31(3). Available from: http://www.orientjchem.org/?p=10887 |
Introduction
Macrocyclic complexes and their cyclic ligands playa major role in expanding the field of bioinorganic chemistry.1Not surprisingly, the number of studies oncomplexes of macrocyclictetraaza ligands involving the synthesis of new ring systems,and the investigation of its function and properties and the discovery of potential applications in the fields of industry, medicine and others,has grown. 2However, there are only a few complexes that involve a neutral macrocyclic ligand [Me6N4H2]with either copper or zinc as the central metal. Examples which have been reported are;[Cu(Me6N4H2)]ClO4,3 [CuI(Me6N4H2)]IH2O,4[CuBr(Me6N4H2)]Br.2H2O,5[ZnI(Me6N4H2)]I3,6[Zn(Me6N4H2)]ClO4,7 7and C18H35Br3N4O2Zn2,8We have reported the preparation of our first copper complex,catena-poly[[5,5,7,12,12,14-hexamethyl-1,4,8,11-tetraazacyclotetradeca-1,7-iene)copper(II)-µ-chlorido[dichlorocuprate(II)-µ-chlorido].9
Previously, most of the macrocyclic complexes were obtained by a template reaction. For example, the complex [(Me6N4H4)·Ni] was derived from the recrystallisation reaction of tris(ethylenediamine) in acetone.10In the present work, we focus primarily on the synthesis of the macrocyclic complex via two separate reactions. Firstly, the synthesis of the protonated macrocyclictetraaza was first performed,followed by the complexation reaction. Since the macrocyclic ligand is protonated (Scheme 1), we may vary its counteranion.11However, the complexation reaction with the protonated form of the tetraazamacrocycleis still not widely reported. Therefore, we have conducted further studies on the complexationreaction using various transition metals.
The complexes of macrocyclictetraaza ligands with various transition metalsplay an importantroleby acting as an active part of metalloenzymes12since the structure of the macrocyclic complexes are similar to the ligand sites of metalloenzymes. Also, both macrocyclic compounds and their complexes have important applications in coordination chemistry and organic catalysis.13,14The complexes can also be used as biomimetic biological model compounds because of their similarities to natural proteins such as hemerythrin and enzymes.15Furthermore, these types of macrocyclic compound are also used as catalysts for biological reactions such as epoxidation or hydrolysis of DNA anddioxygen binding,whilst also acting as active components of antiviral drugs.16 They are also used as medical imaging agents.17Additionally, transition metal macrocyclic complexes have been well tested in tumour treatment methods,radio immunotherapy, and cancer diagnosis.18
In this paper, we report on the complexation of various copper, nickel, and zinc metal salts with the protonated 14-membered ring tetraaza ligand, 5,5,7,12,14,14-hexamethyl-1,4,8,11-tetraazacyclotetradeca-7,14-dienium bromide. The antibacterial properties of both the complexes and the free ligand are also reported.
Experimental
All chemicals and solvents were reagent grade and used as received. C, H and N analyses were determined using Fison model EA 110. The complexes were dried in air. IR spectra (as KBR pellets) were recorded on a Perkin Elmer 400 FT-IR/FT-NIR. Electronic spectra in the 200-800 nm region were recorded on a 1800 Shimadzu spectrophotometer. Magnetic susceptibilities were measure at 25 °C by the Gouy method. The calculation were carried out using μeff = 2.84 √χMcoor.T. The TLC of all the ligand and complexes confirmed their purity.
Synthesis of 14-membered tetraazamacrocyclic ligand
The macrocyclic ligand C16H38N4O2Br2 (4)was synthesised by mixingammonium salt(0.9794 g, 0.01 mol) (1)and ethylenediamine(0.601g, 0.01 mol) (2)in acetone(3) at 80°Cunder constant stirring for 2 h. The solution was then filtered and left overnight at room temperature for crystal growth. Yield: 65%; m.p. 113.4–125.3°C. Anal. Calcd for C16H38N4O2Br2(FW478.32): C,40.2%; H, 7.9%; N, 11.1%. Found: C, 38.9%; H, 7.5%; N, 11.6%. NMR: 1H,δH1.49(6H, s, CH3),δH 2.06 (3H, s, CH3), δH2.80 (2H, s, CH2),δH3.42 (2H, t, CH2),δH3.69 (2H, t, CH2), and δH4.91 (2H, t, NH2+).
Synthesis of (Me6N4H2)Br2.2H2O complexes with copper, nickel, and zinc
The complex was synthesised by stirring together a 1:1 mixture of CuCl2(L(Cu1) and ligand in methanol. The solution was then filtered and left at room temperature for evaporation.The same steps were repeated by changing the ligand to CuBr2(L(Cu2)), Cu(NO3)2(L(Cu3)), CuSO4(L(Cu4)), Ni(OAc)2(L(Ni1)), NiCl2(L(Ni2)), NiSO4(L(Ni3)), Ni(NO3)2(L(Ni4)), ZnCl2(L(Zn1)), Zn(OAc)2(L(Zn2)), ZnSO4(L(Zn3)), and Zn(NO3)2(L(Zn4)). The analytical data are given in Table 1.
Antibacterial Test
The metal salts, free ligands and the complexes formed were tested for antibacterial activity.Three species each of Gram-positive and Gram-negative bacteria were chosen for the test. The complex properties were screened by using the minimum inhibitory concentration (MIC).
Results and Discussion
Characterization of the Ligand
The reaction to synthesise the ligand (4) (Scheme 1)afforded a white crystalline solid in 65% yield. The melting point was 113.4-125.3 °C. Analysis of the microelemental data was in agreement with the expected formula of C16H38N4O2Br2 (experimental: C = 38.9%, H = 7.5%, and N = 11.6%; theoretical: C = 40.2%, H = 7.9%, and N = 11.1%). The FTIR spectrum showed stretching bands at 1667 and 1228 cm-1,indicating the presence of C=N and C-N bonds, respectively. The sharp band at 3468 cm-1 and the broad band at 3012 cm-1 are due to the primary amino and water O-H stretches, respectively. The 1H NMR spectrum confirmed the presence of methyl protons and protonated amine at 1.49, 2.06, and 4.89 ppm, respectively. The signals at 2.80, 3.42, and 3.69 ppm are due to the methylene protons. 13C NMR further confirmed the presence of C=N and C-N bonds with the chemical shifts of 175.87 and 58.35 ppm, respectively. TheUV-VIS spectrum showed a single absorption peakat 238 nm, which may be assigned as the π → π* electronic transition of the C=Nchromophore.
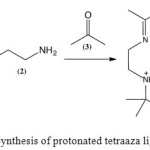 |
Scheme 1: Synthesis of protonated tetraaza ligand Click here to View scheme |
Characterization of Metal Complexes
A suspension of ligand (L) in methanol at reflux reacts with metal(II) salts to generate the corresponding complexes (Scheme 2) in moderate yield (49-88%). Analytical and physical data (Table 1) and spectral data (Table 2 and 3) showed that some of the complexes are compatible with the proposed structures. The complexes are coloured, stable in air partially soluble in common solvents.
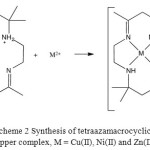 |
Scheme 2: Synthesis of tetraazamacrocyclic copper complex, M = Cu(II), Ni(II) and Zn(II) |
The elemental analysis data for all complexes except L(Ni2), L(Ni3), and L(Ni4)shows a high level of agreement with its theoretical value. However, the percentages of N atom for the complexes are quite different than the theoretical values. This abnormality was due to faulty detector for N atom at the time of measurement. Vibrations for N-H (amine), C-H (methyl), C-N (imine), and C=N (azomethine) in IR spectra are the most important vibrations that each complex must contain. The presence of these indicates that the formed complexes may contain the original ring structure of the tetraazamacrocycle. Table 2 shows the FTIR frequencies of the complexes. The absence of C=N (azomethine) vibrations in L(Ni2), L(Ni3), and L(Ni4), indicates that their complexes may have different structures and that the tetraazamacrocycle may have undergone a ring opening.Table 3 shows the UV-VIS and magnetic moments (µeff) data for all the complexes formed. L(Zn1), L(Zn2), L(Zn3), L(Zn4) and L(Ni2)do not show µeff effects or d-d transitions because all the zinc complexes are diamagnetic; the d-orbital (d10) of zincis already full and no other possible transitions may occur. The absence of any d-d transition in L(Ni4), indicates that the product formed does not contain any nickel.
Table 1: Elemental analysis and melting point data
|
Complexes |
Colour |
Yield (%) |
Experimental (Theoretical) |
Melting point (ºC) |
||
|
C |
H |
N |
||||
|
L(Cu1) |
Purple |
65 |
31.0(31.0) |
5.0(6.8) |
11.4(9.0) |
208.0–208.3 |
|
L(Cu2) |
Purple |
88 |
28.2 (28.7) |
5.1 (5.7) |
7.8 (8.4) |
225.4–225.8 |
|
L(Cu3) |
Purple |
69 |
26.5 (26.7) |
4.7 (5.0) |
7.6 (7.8) |
217.6–218.0 |
|
L(Cu4) |
Purple |
70 |
28.2 (28.9) |
7.0 (7.2) |
9.5 (8.5) |
199.6 – 200.4 |
|
L(Ni1) |
Yellow |
73 |
36.0 (36.1) |
6.3 (6.4) |
14.9 (10.5) |
328.6 – 332.1 |
|
L(Ni2) |
Orange |
65 |
13.0 (26.9) |
4.7 (4.5) |
24.2 (7.8) |
319.7 – 325.0 |
|
L(Ni3) |
Green |
55 |
9.7 (25.9) |
5.2 (4.2) |
14.4 (7.8) |
328.0 – 328.4 |
|
L(Ni4) |
Green |
49 |
6.1 (25.9) |
5.0 (4.4) |
7.2 (7.6) |
327.9 – 331.0 |
|
L(Zn1) |
Yellow |
70 |
30.3 (31.3) |
6.1 (6.2) |
15.2 (9.1) |
198.0 – 200.0 |
|
L(Zn2) |
White |
60 |
35.9 (36.7) |
6.5 (6.5) |
7.3 (10.7) |
260.0 – 265.0 |
|
L(Zn3) |
White |
64 |
29.9 (30.8) |
5.5 (5.9) |
7.2 (10.7) |
240.0 – 243.0 |
|
L(Zn4) |
White |
68 |
30.3 (30.7) |
9.1 (9.3) |
4.2 (11.1) |
182.0 – 199.8 |
Table 2: FTIR stretching frequencies for the complexes
|
Complexes |
Stretching frequencies (cm-1) |
|||
|
N-H (amine) |
C-H (methyl) |
C-N (imine) |
C=N (azomethine) |
|
|
L(Cu1) |
3158.4 |
2973.9 |
1169.7 |
1662.9 |
|
L(Cu2) |
3148.8 |
2969.8 |
1164.4 |
1670.8 |
|
L(Cu3) |
3158.7 |
2973.6 |
1169.3 |
1662.6 |
|
L(Cu4) |
3424.4 |
2968.7 |
1171.4 |
1632.7 |
|
L(Ni1) |
3424.7 |
2868.5 |
1165.7 |
1653.9 |
|
L(Ni2) |
3175.2 |
2919.1 |
1033.5 |
– |
|
L(Ni3) |
3168.9 |
2921.9 |
1030.7 |
– |
|
L(Ni4) |
3170.5 |
2919.1 |
1034.7 |
– |
|
L(Zn1) |
3200.0 |
2964.0 |
1033.0 |
1656.0 |
|
L(Zn2) |
3211.2 |
2961.9 |
1107.9 |
1660.3 |
|
L(Zn3) |
3202.9 |
2968.3 |
1664.1 |
1159.6 |
|
L(Zn4) |
3118.1 |
2967.6 |
1084.1 |
1604.2 |
Table 3: UV-VIS and µeffdata for the synthesised complexes
|
Complexes |
µeff |
Transition π → π* |
Transition d-d |
|
L(Cu1) |
2.45 |
246.0 |
519.0 |
|
L(Cu2) |
2.31 |
237.5 |
520.5 |
|
L(Cu3) |
– |
242.5 |
516.5 |
|
L(Cu4) |
2.01 |
234.0 |
519.0 |
|
L(Ni1) |
0.87 |
241.5 |
436.5 |
|
L(Ni2) |
– |
276.0 |
– |
|
L(Ni3) |
– |
271.5 |
421.0 |
|
L(Ni4) |
1.39 |
277.0 |
– |
|
L(Zn1) |
– |
247.0 |
– |
|
L(Zn2) |
– |
247.0 |
– |
|
L(Zn3) |
– |
220.0 |
– |
|
L(Zn4) |
– |
250.0 |
– |
X-ray crystallography
Although all the products were formed as solid crystals, not all were suitable for X-ray crystallographic analysis. The crystals of the complexes formed were analysed using Bruker APEX-II CCD fitted with Mo Kα radiation. Only copper tetraazamacrocylic complex (L(Cu1)) and nickel (II) tetraazamacrocylic complex (L(Ni1))were suitable for X-ray analysis. The X-ray structure of each product complemented the results obtained by elemental analysis, FTIR and UV spectroscopy.21The structural data for the complex of L(Cu1)(Figure 1) and L(Ni1)(Figure 2) showed good agreement withits elemental analysis data. Moreover, its FTIR proved that all the important vibrations were present. The UV-VIS analysis also showed the presence of the azomethinechromophore and d-d transition, strongly indicating that the product maintained its original tetraaza ligand structure.However, the structure of several related macrocyclic complexes have been reported for many times in the past years using different salt of metal (II) ions. The experimental details for the molecular structure and structure refinement data for the complexes of L(Cu1)and L(Ni1)are shown in Table 4. Selected bond distance and bond angles are listed in Table 5.
Table 4: Crystal data and refinement parameters for L(Cu1) and L(Ni1) complexes
|
L(Cu1) |
L(Ni1) |
|
| Empirical formula |
C16H36Br2CuN4O2 |
C16H34Br2N4NiO2 |
| Formula weight |
539.85 |
533.00 |
| Crystal system |
Monoclinic |
Monoclinic |
| Space group |
P 21/c |
P 21/c |
| A (Å) |
8.0453(8) |
8.0402(5) |
| B (Å) |
15.6095(19) |
15.6070(10) |
| c (Å) |
8.9229(10) |
8.9200(6) |
| Α (°) |
90 |
90 |
| Β (°) |
99.844(4) |
99.800(2) |
| γ (°) |
90 |
90 |
| V(Å) |
1104.1(2) |
1102.98(12) |
| Z |
2 |
2 |
| F(000) |
550 |
544 |
| Density (mg/m3) |
1.624 |
1.605 |
| Absorption coefficient, μ (mm-1) |
4.625 |
4.518 |
| Crystal size (mm) |
0.300 x 0.280 x 0.250 |
0.500 x 0.160 x 0.150 |
| θrange for data collection (°) |
2.882 to 25.496° |
3.415 to 25.999 |
| Goodness-of-fit on F2 |
1.106 |
1.089 |
| Independent reflections/Rint |
2055/0.1379 |
2155/0.0767 |
| Data / restraints / parameters |
2055 / 3 / 130 |
2155 / 0 / 126 |
| R1 [I> 2σ(I)] |
0.0485 |
0.0409 |
| wR2 [I> 2σ(I)] |
0.1017 |
0.0913 |
Table 4B: Results of MIC tests using the tetraaza ligand, complexes and metal salts
|
Bacteria |
Complex/Metal Salts |
||||||
|
TBr |
L(Cu1) |
L(Ni1) |
L(Zn1) |
CuCl2 |
Ni(OAc)2 |
ZnCl2 |
|
|
Staphylococcus aureus |
– |
6.3 |
25.0 |
3.1 |
1.6 |
– |
0.9 |
|
Bacillus subtilis |
25.0 |
3.1 |
25.0 |
25.0 |
0.8 |
1.6 |
3.1 |
|
Enterococcus faecalis |
– |
6.3 |
25.0 |
12.5 |
– |
1.6 |
1.6 |
|
Enterobacteraerogenes |
– |
3.1 |
25.0 |
25.0 |
0.8 |
6.3 |
3.1 |
|
Pseudomonas aeruginosa |
25.0 |
6.3 |
25.0 |
25.0 |
– |
3.1 |
6.3 |
|
Escherichia coli |
12.5 |
6.3 |
25.0 |
12.5 |
0.8 |
1.6 |
1.6 |
Table 5: Selected bond length and angle for L(Cu1)and L(Ni1)
|
Bond Lengths (Å) |
L(Cu1) |
Bond Lengths (Å) |
L(Ni1) |
|
Cu1-N1 |
1.904(5) |
Ni1-N1 |
1.936(3) |
|
M1-N2 |
1.929(5) |
Ni1-N2 |
1.919(3) |
|
N1-C5 |
1.280(8) |
N1-C6 |
1.499(5) |
|
N1-C1 |
1.477(8) |
N1-C2 |
1.482(5) |
|
N2-C2 |
1.465(9) |
N2-C4 |
1.276(5) |
|
N2-C3 |
1.496(8) |
N2-C3 |
1.480(5) |
|
Bond Angle (°) |
Bond Angle (°) |
||
|
N1-Cu1-N2 |
86.02(2) |
N2-Ni-N1 |
86.02(13) |
|
C1-N1-Cu1 |
111.5(4) |
C3-N2-Ni1 |
111.7(2) |
|
C2-N2-Cu1 |
108.1(4) |
C2-N1-Ni1 |
107.0(2) |
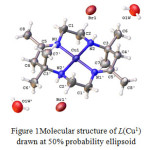 |
Figure 1: Molecular structure of L(Cu1)drawn at 50% probability ellipsoid |
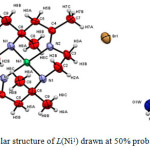 |
Figure 2: Molecular structure of L(Ni1) drawn at 50% probability ellipsoid |
Antibacterial studies of the metal salts,synthesised tetraazaligand and complexes
The free ligand and its complexes were tested as antibacterial agents with the help of MIC tests. The compounds were dissolved in distilled water with an initial concentration of 25mg/mL.Three Gram-positive bacteria (Staphylococcus aureus, Bacillus subtilis,and Enterococcus faecalis)and three Gram-negative bacteria(Escherichia coli,Enterobacteraerogenes,and Pseudomonas aeruginosa)were selected for the test. The bacterial single colony was maintained in nutrient agar and was subcultured in Mueller-Hinton broth for the tests. The solution was diluted eight times, and positive and negative controls were duplicated three times for comparison using a 96 well plate. The results are tabulated in Table 4.
Each macrocyclic complex exhibited higher antibacterial activity than its free ligand. The complexes and its free ligand (L) have similar structures, but the complex shows one difference in that it contains a metal ion that is bonded directly to four nitrogen atoms in the ring. The antibacterial activity of the complexes can be explained with the help of chelating reactions. Chelates reduce the polarity of the metal ion because of the overlapping of the ligand orbital and the sharing of the positive charge between the metal and the electron-donating group. Therefore, the complexes are much more stable than the free ligands due to the delocalisation of π electrons. This helps the complexes in penetrating the lipid membrane and blocking the metal binding site on the enzyme within the microorganism.22 In addition, the copper complex of tetraazamacrocycleL(Cu1) also blocks the cell respiration process, preventing the synthesis of protein and therefore inhibiting the growth of the organism.23The MIC value for the metal salts used for the complexation reactions ofL(Cu1), L(Ni1), and L(Zn1) showed better results than their complexes. This is because the size of the metal salt molecule is much smaller than that of the complex,making it easier for the salt to penetrate the membrane cells.
Conclusion
Complexation reactions of tetraazamacrocyclic ligand with copper, nickel, and zinc resulted in the formation of new complexes. The complexationreaction of L(Cu1)and L(Ni1)involved the deprotonation of the amino nitrogen atoms, but with the same bromide counteranion from the ligand. The complexation reaction of L(Ni3) and L(Ni4) showed that opening of the tetraazaring occurred. L(Cu1), L(Ni1)and L(Zn1) showed antimicrobial activity to the selected bacteria.
Acknowledgement
A.A.S.H. would like to thank the Universiti Kebangsaan Malaysia (UKM) for the MSc Scholarship (Zamalah, UKM). S.F.M.Y would like to thank MOSTI and the Universiti Kebangsaan Malaysia for the research grants (DLP-2013-009 and DIP-2014-16).
References
- El-Tabl. A.S. Bull. Korean Chem. Soc.2004, 25, 1757-1761
- Sujatha, S., Balasubramanian, S., Fun, H.K., and Chinnakali, K.Polyhedron 2008, 27, 1925-1929
- Lu, T.H., Lee, T.J., Liang, B.F., and Chung, C.S. J. Inorg. Nucl. Chem.1981,43, 2333-2336
- Podberezskaya, N.V., Pervuknina, N.V., and Myachina, L.I. J. Struct. Chem.1986, 27,268-272
- Shi, F.F., and He, X.L. Acta Crystallogr. Sect. E: Struct. Rep. Online, 2011, 67, 607
- Heinlein, T., and Tebbe, K.F. Z. Kristallogr.1985,170, 170-171
- Yang, Y.M. Acta Crystallogr. Sect. E: Struct. Rep. Online, 2005, E61, m1618-m1619
- Li, H.F., Shi, F.F., and Rong, R. Acta Crystallogr. Sect. E: Struct. Rep. Online, 2010, E66, 1338
- Yamin, B.M., Ismail, W., and Daran, J.C. ActaCrystallogr. Sect. E: Struct. Rep. Online, 2012, E68, 886-887
- Curtis, N.F., and House, D.A. Chemistry and Industry 1961,6, 1708-1709
- Mohamad Wafiuddin Haji Ismail. Sintesisgaram 5,5,7,12,12,14-heksametil-1,4,8,11-tetraazasiklotetradeka-7,14-dienium, Pengkompleks and enganLogam&AktivitiBiologi. Master Thesis, Graduate Studies Centre, UKM. (2012)
- Chaudhary, A., Dave, S., Swaroop, R., and Singh, R.V. J. Indian Chem. Soc. 2002, 79, 371-373
- Ciampolini, M., Nardi, N., Vantacoli, B., and Micheloni, M. Coord. Chem. Rev. 1992, 120, 223-236
- Li, Z., Ding, H., and Wu, S. Mater. Res. Bull.2013, 48: 1920-1926
- Najlaa al-Radadi, S., Sawsan Al-Ashqar, M., and Mohsen Mostafa, M. J. Incl. Phenom. Macro. Chem.2011, 69, 157-165
- Frausto da Silva, J.J.R., and Williams, R.J.P. The Biological Chemistry of the Elements, Clarendon Press, Oxford. (1991)
- Lauffer, R.B. Chem. Rev.1987, 87, 901-927
- Norman, T.J., Parker, D., Smith, F.C., and King, D.J. Supramol. Chem.1995, 4, 305-308
- Rohovec, J., Vojtisek, P., and Lukes, I. Collection of Czechoslovak Chemical Communications1999, V64, 73-88
- Kennedy, A.R., Lutta. S.T., Morisson, C.A., Okoth, M.O., and Orang’o, D.M. ActaCrystallogr. Sect. E: Struct. Rep. Online, 2011, E67,682-683
- Hassan, N.H., Ali, N.M., Yamin, B.M., AbdKarim, N.H. and AbdGhani, N.A. The Malaysian J. Anal. Sci.2014, 18, 562-571
- Prayong, P., Barusrux, S.,and Weerapreeyakul, N. Fitoterapia2008. 79, 598-601
- Sharma, R., Prabhat, Singh,.R.,Pawar, S. and Chauhan, A. J. Am. Sci.2010, 6, 559-564

This work is licensed under a Creative Commons Attribution 4.0 International License.









