Microbial Transformation of (-)-Alloisolongifolene
Naik T. Khan1, Muhammad Atif1 and Amal Al-Aboudi2*
1 H. E. J. Research Institute of Chemistry, International Center for Chemical and Biological Sciences, University of Karachi, Karachi-75270, Pakistan.
2Chemistry Department, The University of Jordan, Amman11942, Jordan
DOI : http://dx.doi.org/10.13005/ojc/300304
Article Received on :
Article Accepted on :
Article Published : 23 Aug 2014
(-)-Alloisolongifolene (1) is a structural analogue of the naturally occurring longifolene. The microbial transformation of (-)-alloisolongifolene (1) by Cunninghamella elegans afforded two new metabolites, 3a-hydroxyalloisolongifolol (2) and 13-hydroxyalloisolongifolol (3). The structures of the metabolites were deduced on the basis of their spectral data, including 1D and 2D NMR, IR and HREIMS.
KEYWORDS:(-)-Alloisolongifolene; Microbial Transformation; Cunninghamella elegans; 3-Hydroxyalloisolongifolol; 13-Hydroxyalloisolongifolol
Download this article as:| Copy the following to cite this article: Khan N. T, Atif M, Aboudi A. A. Microbial Transformation of (-)-Alloisolongifolene. Orient J Chem 2014;30(3). |
| Copy the following to cite this URL: Khan N. T, Atif M, Aboudi A. A. Microbial Transformation of (-)-Alloisolongifolene. Orient J Chem 2014;30(3). Available from: http://www.orientjchem.org/?p=4300 |
INTRODUCTION
Microbial transformation of terpenes has been an attractive method to produce new metabolites with enhanced biological activity1. Insertion of a hydroxyl group in one step, with high regio- and stereo-selectivity, at an inactive terpenoidal skeleton by microbial transformation remains unmatched by chemical means 2. Hydroxylation of terpenes is of importance to the flavor and fragrance industry in the search for new production methods and new compounds 3. A large number of sesquiterpenes with a bridged tricyclic skeleton have been isolated from plants and fungi 4, some of them have exhibited interesting biological properties 5. However, only a few reports about their fungal transformation have appeared 6. Recently, biotransformation of various bridged sesquiterpenes has been described by Choudhary et al 5, 7, 8, who reported regio- and stereo-selective hydroxylation of (+)-cycloisolongifol-5b-ol , (-)-isolongifolol and (+)-isolongifolene-4-one by different fungi species.
(-)-Alloisolongifolene (1), a bridged tricyclic sesquiterpene, produced by acid-catalyzed isomerization of the naturally occurring longifolene 9, 10.
Biotransformation of 1 has been reported by using enzymes from chicory roots (Cichorium intybus) where the hydroxylation took place at an isopropylidene group at the allylic position 11.
In the present study, we report that the microbial transformation of (-)-alloisolongifolene (1) using Cunninghamella elegans afforded two new hydroxylated metabolites (2 and 3).
MATERIAL AND METHODS
General experimental procedures
IR Spectra were recorded in CHCl3 on an FTIR-8900 spectrophotometer. Optical rotations were measured on a JASCO DIP-360 digital polarimeter. UV Spectra were recorded in CHCl3 solution on a Hitachi U-3200 spectrophotometer. The 1H and 13C NMR spectra were recorded in CDCl3 solution on a Bruker Avance-400 NMR at 400 and 100 MHz, respectively (d ppm, J in Hz). The EI and HREI MS were recorded on JEOL JMS 600H mass spectrometer. TLC was performed using precoated plates (Silica gel 60, PF254, 0.2 mm, Merck). Compound 1 was purchased from Fluka.
Organism and media
Microbial culture, Cunninghamella elegans (NRRL 1392), was obtained from Northern Regional Research Laboratories (NRRL) and maintained on SDA and stored at 4 oC prior to use. The fermentation medium for Cunninghamella elegans (NRRL 1392), was prepared by dissolving the following ingredients in distilled water (3.0 L): glucose (30.0 g), glycerol (30.0 mL), peptone (15.0 g), yeast extract (15.0 g), KH2PO4 (15.0 g), and NaCl (15.0 g).
General fermentation and extraction procedure
The fermentation media thus obtained was distributed among 30 flasks of 250 mL capacity (100 mL each) and autoclaved. The fermentation was carried out according to a standard stage II protocol 12, 13. Compound 1 (600 mg) was dissolved in 15 mL acetone and the solution was evenly distributed among the 30 flasks (20 mg/ 0.5 mL in each flask) containing 24 h culture of Cunninghamella elegans together with control flasks. Fermentation continued further for an additional 3 days on a rotatory shaker (200 rpm) at 29° C. During the fermentation, aliquots from flasks were taken daily and analyzed by TLC in order to determine the degree of transformation of the substrate. One control flask, without biomass (for checking substrate stability), and one flask without exogenous substrate (for checking endogenous metabolite) were used. The culture media and mycelium were separated by filtration. The mycelium was washed with CH2Cl2 (1.0 L) and the filtrate was extracted with CH2Cl2 (3 ´ 3 L). The combined organic extracts was washed with brine, dried over anhydrous Na2SO4 and then evaporated under reduced pressure to yield a crude brown gum (2.6 g). Control flasks were also harvested and their contents tested by TLC to detect any biotransformed products. The crude residue (2.6 g) was subjected to column chromatography. Elution with petroleum ether and EtOAc yielded compounds 2 and 3 in 4.2%, and 2.8% yields, respectively.
3a-Hydroxyalloisolongifolol (2) was obtained as an amorphous material
[a]25D: +68 (c = 0.4, CHCl3). UV (CHCl3): λmax (logε) = 202 nm (1.8). IR (CHCl3): νmax = 3419, 2992 cm-1. 1H NMR (CDCl3, 400 MHz): see Table 1. 13C NMR (CDCl3, 100 MHz): see Table 2. MS (EI, 70 eV): m/z (%): 238 (7) [M+], 220 (14), 206 (6), 166 (38), 146 (28), 120 (100), 78 (74), 55 (28). MS (HREI): m/z = 238.1084 (C15H26O2, calcd 238.1093).
13-Hydroxyalloisolongifolol (3) was obtained as an amorphous material
[a]25D: +116 (c = 0.5, CHCl3). UV (CHCl3): λmax (logε) = 203 nm (2.2). IR (CHCl3): νmax = 3401, 2986 cm-1. 1H NMR (CDCl3, 400 MHz): see Table 1. 13C NMR (CDCl3, 100 MHz): see Table 2. MS (EI, 70 eV): m/z (%) = 238 (16) [M+], 220 (31), 208 (26), 166 (26), 152 (16), 120 (100), 78 (28), 55 (47). MS (HREI): m/z = 238.1074 (C15H26O2, calcd 238.1086).
RESULTS AND DISCUSSION
To investigate the capacity of Cunninghamella elegans to metabolise compound 1, a small-scale experiment was conducted. TLC of the crude chloroform extract indicated the presence of two new more polar metabolites. Subsequently, a large-scale fermentation was carried out to produce sufficient quantities of these metabolites for structure elucidation. Biotransformation of compound 1 with Cunninghamella elegans for 3 days yielded the new metabolites 2 and 3 (Scheme-1).
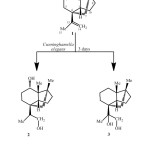 |
Scheme1: Biotransformation of (-)-alloisolongifolene (1) with Cunninghamella elegans.Click here to View Scheme |
The HREIMS of compound 2 exhibited M+ at m/z 238.1084, corresponding to the formula C15H26O2 (calcd 238.1093), which is 34 a.m.u. higher than the substrate 1. The IR spectrum of compound 2 showed absorption at 3524 cm-1 indicating the presence of hydroxyl groups. The 1H NMR spectrum (Table 1) of compound 2 was quite different from the parent compound 1. The olefinic proton signals at d 4.73 and 4.63 of 1 were absent, while three new additional signals appeared at 83.57 (1H, dd, J3β,4a = 15.1 Hz, , J3β,4β = 6.0 Hz), 3.60 (1H, dd, J14a,14β = 12.2 Hz, J14,13 = 6.0 Hz) and 3.38 (1H, t, J = 12.2 Hz), assigned for H-3 and the two vicinal protons attached to C-14, respectively. The 13C-NMR and DEPT spectra of 2 (Table 2) confirmed anti-Markovnikove’s hydration of the double bond. It showed the absence of the double bond signals, the presence of a CH at 847.5 and a downfield CH2 at 866.5, assigned to C-13 and C-14, respectively. The downfield CH signal at d 72.1, the absence of the methylene carbon at d 38.8 and the downfield shifts of C-2 and C-4 signals (Table 2) indicated hydroxylation at C-3 of compound 2. COSY 45° and HMBC experiments supported these assignments. COSY 45° showed correlations between H-14 (83.60, 3.38) and H-13 (1.76), and HMBC showed correlations between H-14 (83.60, 3.38) and C-15 (815.9), and C-13 (847.5). The position of the second OH group at C-3 was confirmed by the HMBC correlations of H-3 (83.57) with C-12 (815.4), C-4 (847.8), and C-2 (853.2). The stereochemistry of the C-3 hydroxyl group was determined as a on the basis of NOESY correlations between H-3 (83.57) and each of H-1β (81.80), H-5β (81.62) and Me-12β (80.94). The structure of the new metabolite 2 was thus established as 3a-hydroxyalloisolongifolol.
Table 1. 1H NMR chemical shift data of the compounds 1-3.
| Position | 18H (J in Hz) | 28H (J in Hz) | 38H (J in Hz) |
| 1 | 1.52, m | 1.80, m | 2.05, m |
| 2 | – | – | – |
| 3 | 1.44; 1.12, m | 3.57 (dd, J = 15.1, 6.0 Hz) | 1.30; 1.18, m |
| 4 | 1.57; 1.49, m | 1.67; 1.04, m | 1.54; 1.44, m |
| 5 | 1.60; 1.40, m | 1.62; 1.12, m | 1.69; 1.37, m |
| 6 | – | – | – |
| 7 | 1.53; 1.35, m | 1.58; 0.82, m | 1.50; 1.35, m |
| 8 | – | – | – |
| 9 | 1.55; 1.47, m | 1.59; 1.25, m | 1.50; 1.44, m |
| 10 | 1.59; 1.22, m | 1.56; 0.97, m | 1.48; 1.31, m |
| 11 | 0.76, s | 0.86, s | 0.80, s |
| 12 | 0.87, s | 0.94, s | 0.87, s |
| 13 | – | 1.76, m | – |
| 14 | 4.73; 4.63 (br s) | 3.60 (dd, J = 12.2, 6.0 Hz); 3.38 (t, J = 12.2, Hz) | 4.03; 3.26 (d, J = 10.7 Hz) |
| 15 | 1.67, s | 1.01 (d, J = 6.8 Hz) | 1.09, s |
The HREI MS of metabolite 3 showed the M+ at m/z 238.1074 (C15H26O2, calcd 238.1086), similar to that of compound 2. The 1H NMR spectrum (Table 1) showed AB doublets at d 4.03 (J14a,14b = 10.7 Hz) and 3.26 (J14b,14a = 10.7 Hz), indicating the hydroxylation of the C-13/C-14 double-bond. The 13C NMR spectrum (Table 2) of compound 3 was similar to that of compound 1, except for the absence of the double bond and the appearance of additional downfield OH bearing signals at d 71.6 (C-14), and 83.8 (C-13). The dihydroxylation of the double bond was further confirmed by the HMBC correlations between Me-15 (d 1.09) and C-13 (d 83.8), C-14 (d 71.6), and C-6 (d 51.9). Based on the above mentioned spectral data, the structure of the new metabolite 3 was deduced as 13-hydroxyalloisolongifolol.
Table 2. 13C NMR Chemical shift data of the compounds 1-3.
| Position | 18C | 28C | 38C |
| 1 | 51.8, d | 41.8, d | 50.9, d |
| 2 | 46.8, s | 53.2, s | 46.7, s |
| 3 | 38.8, t | 72.1, d | 38.5, t |
| 4 | 20.7, t | 47.8, t | 22.2, t |
| 5 | 44.0, t | 39.0, t | 44.1, t |
| 6 | 49.7, s | 46.5, s | 51.9, s |
| 7 | 22.0, t | 19.8, t | 21.6, t |
| 8 | 47.9, s | 44.2, s | 46.9, s |
| 9 | 32.5, t | 29.2, t | 35.2, t |
| 10 | 36.5, t | 23.9, t | 36.2, t |
| 11 | 19.8, q | 12.5, q | 15.6, q |
| 12 | 15.4, q | 15.4, q | 22.9, q |
| 13 | 152.5, s | 47.5, d | 83.8, s |
| 14 | 109.4, t | 66.5, t | 71.6, t |
| 15 | 20.5, q | 15.9, q | 20.1, q |
It has been reported that the filamentous fungus Cunninghamella elegans contains enzymes with activities similar to those found in mammals, some of which are P-450 mono-oxygenase and epoxide hydrolase 14. So we think that compound 2 may have formed from anti hydrolysis of the corresponding epoxide as shown in scheme 2.
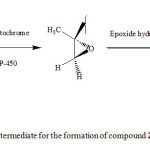 |
Scheme 2: Proposed intermediate for the formation of compound 2. Click here to View Scheme |
Cunninghamella elegans fungus was able to bio-catalyze the hydroxylation of the bridged sesquiterpenes, (-)-alloisolongifolene (1), in a regio and stereo-specific manner. It introduced a hydroxyl group at the 3a-position, and hydrate the double bond in an anti-Markovnikov’s producing a new compound 3a-Hydroxyalloisolongifolol (2), in addition to oxidation of the double bond (C-13, C-14) to the corresponding diol to give the new compound 13-Hydroxyalloisolongifolol (3).
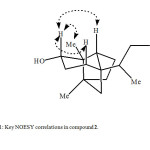 |
Fig1: Key NOESY correlations in compound 2.Click here to View Figure |
ACKNOWLEDGEMENTS
The authors acknowledge the Higher Education Commission, Islamabad, Pakistan for the financial support through Indigenous Ph. D. Fellowship Program, and acknowledge the assistance of Prof. Atta-ur-Rahman and Prof. Mohammad Iqbal Choudhary though out this work.
REFERENCES
- Bhatti, H. N.; Zubair, M.; Rasool, N.; Hassan, Z.; Ahmad, V. U. Nat. Prod. Commun. 2009. 4(8), 1155-1168.
- Rocha, B. A.; Pupo, M. T.; Antonucci, G. A.; Sampaio, S. V.; Paiva, R. M. V.; Said, S.; Gobbo-Neto, L.; Costa, F. B. J. Ind. Microbiol. Biotechnol. 2012, 39(11), 1719-1724.
- Faber, K. Biotransformations in organic chemistry: A Textbook. 4th ed. Springer: Berlin, (2000).
- Cohen, H.; Charrier, C.; Ricard, L.; Perreau, M. J. Nat. Prod. 1992, 55, 326-332.
- Choudhary, M. I.; Musharraf, S. G.; Ahmad, N. S.; Anjum, S; Parvez, M.; Fun, H. K.; Atta-ur-Rahman. Bioorg. Med. Chem. 2005, 13(6), 1939-1944.
- Aleu, J.; Hanson, J.; R.; Galán, R. H.; Collado, I. G. J. Nat. Prod. 1999, 62, 437-440.
- Choudhary, M. I.; Kausar, W.; Siddiqui, Z. A.; Atta-ur-Rahman. Z. Naturforsch. 2006, 61b, 1035–1038.
- Choudhary, M. I.; Musharraf, S. G.; Khan, M. T. H.; Abdelrahman, D.; Parvez, M.; Shaheen, F.; Atta-ur-Rahman. Helv. Chim. Acta 2003, 86(10), 3450–3460.
- Shitole, H. R.; Pramod, V.; Nayak, U. R. Tetrahedron Lett. 1983, 24(23), 2411–2412.
- Mota, J. S.; Souza, D. S.; Boone, C. V.; Cardoso, C.A. L.; Caramao, E. B. J. Essent. Oil Bear. Pls. 2013, 16(1), 11-16.
- Kraker, D.; Schurink, M.; Franssen, M. C. R.; König, W. A.; Groot, A.; Bouwmeester, H. J. Tetrahedron 2003, 59(3),409-418.
- Betts, R. E.; Walters, D. E.; Rosazza, J. P. J. Med. Chem. 1974, 17(6), 599–602.
- Choudhary, M. I.; Nasir, M. Khan, S. N.; Atif, M.; Ali, R. A. Khalil, S. M.; Atta-ur-Rahman. Z. Naturforsch. 2007, 62b, 593 – 599.
- Wackett, L. P.; Gibson, D. T. Biochem. J. 1982, 205, 117–122.

This work is licensed under a Creative Commons Attribution 4.0 International License.









