Geometrical Aspects of Trinuclear Tetra-µ-Chloro-Cadmium Complex with 5-Methyl-4H-1,2,4-Triazole-3,4-Diamine
Mohammad Hakimi*, Zahra Mardani and Keyvan Moeini
Chemistry Department, Payame Noor University, 19395-4697 Tehran, I.R. Iran.
Article Received on :
Article Accepted on :
Article Published : 19 Oct 2016
In this work, a cadmium complex [Cd3(L)2(HL)2(µ-Cl)4Cl4] with the ligand L, 5-methyl-4H-1,2,4-triazole-3,4-diamine,was prepared and identified by elemental analysis, FT-IR spectroscopy and single-crystal X-Ray diffraction. The cadmium atom in the crystal structure of 1 has distorted octahedral geometry by coordination of the two nitrogen atom of L and four chloride ions. Two chloride and a N–N bridges connect two adjacent cadmium atoms.
KEYWORDS:Trinuclear; Cadmium Complex; Triazole; X-ray Crystal Structure
Download this article as:| Copy the following to cite this article: Hakimi M, Mardani Z, Moeini K. Geometrical Aspects of Trinuclear Tetra-µ-Chloro-Cadmium Complex with 5-Methyl-4H-1,2,4-Triazole-3,4-Diamine. Orient J Chem 2013;29(1). |
| Copy the following to cite this URL: Hakimi M, Mardani Z, Moeini K. Geometrical Aspects of Trinuclear Tetra-µ-Chloro-Cadmium Complex with 5-Methyl-4H-1,2,4-Triazole-3,4-Diamine. Available from: http://www.orientjchem.org/?p=22577 |
Introduction
Amongst five-membered ring systems available, the presence of the three nitrogens in triazoles provides an interesting class of compounds. Triazoles in particular, substituted-1,2,4-triazole are among various heterocycles that have received the most attention during last two decades as potential antimicrobial agents, antifungal, antitubercular, anti-HIV, antiinflammatory, CNS stimulants, sedatives, antianxiety1. A larger variety of 1,2,4-triazole-based ligands have been used for preparation of polymeric coordination networks2 and synthesis of nitrogen-rich energetic materials3.
In the past few years we have studied complexation of multi N-donor ligands4−7. In continuation of our previous studies, in this work the preparation of cadmium(II) complex, [Cd3(L)2(HL)2(μ-Cl)4Cl4] (1), with 5-methyl-4H-1,2,4-triazole-3,4-diamine (L, scheme 1) was described. This complex was characterized by elemental analysis, IR spectroscopy and X-ray crystallography.
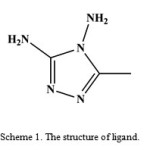 |
Scheme 1: The structure of ligand. |
Material and Methods
General methods
All starting chemicals and solvents were reagent or analytical grade and used as received. The infrared spectrum of a KBr pellet was recorded in the range 4000–400 cm−1 using a FT-IR 8400-Shimadzu spectrometer. The carbon, hydrogen and nitrogen contents were determined in a Thermo Finnigan Flash Elemental Analyzer 1112 EA. Melting point was determined using a Barnsted Electrothermal 9200 electrically heated apparatus.
Synthesis of [Cd3(L)2(HL)2(μ-Cl)4Cl4], 1.
A solution of L. HCl (1.3 mmol, 0.19 g), dissolved in 10 mL EtOH, were added to a 10 mL ethanolic solution of CdI2 (1 mmol, 0.36 g). The reaction mixture was stirred at 60 °C for 2 h and a white precipitate was formed and then filtrated. The resulting solution was left at room temperature for several days and colorless crystals suitable for X-ray diffraction were collected. The results of melting point, CHN analysis and IR spectroscopy of the crystals were identical to those of the initial precipitate. Yield (0.25 g) 83%; Decomposed > 270 °C. Anal. Calcd for C12H30Cd3Cl8N20 (%): C, 13.40; H, 2.81; N, 26.05. Found: C, 13.42; H, 2.81; N, 25.88. IR (KBr, cm−1): 3317 s (νas NH2), 3240 m (νs NH2), 3183 m (ν NH), 1697 s (ν C=N), 1643 m (δ NH2),1435 w (δas CH3), 1381 w (δs CH3), 1281 w (ν CN), 1092 w (ν NN).
Crystal structure determination and refinement
Intensity data were collected on a Bruker APEX-II CCD area detector diffractometer with graphite monochromated Mo Ka radiation (50 kV, 30 mA) using the APEX-II8 data collection software. The collection method involved w-scans of width 0.5° and 512 × 512 bit data frames. Data reduction was carried out using the program SAINT+9 and face indexed absorption corrections were made using the program XPREP9. The crystal structure was solved by direct methods using SHELXTL10. Non-hydrogen atoms were first refined isotropically followed by anisotropic refinement by full matrix least-squares calculations based on F2 using SHELXTL. Hydrogen atoms were first located in the difference map then positioned geometrically and allowed to ride on their respective parent atoms. Crystallographic data and details of the data collection and structure refinement are listed in table 1. Selected bond lengths and angles are listed in table 2.
Table 1. Crystal data and structure refinement for 1.
| Empirical formula | C12H30Cd3Cl8N20 |
| Formula weight (g mol−1) | 1075.36 |
| Temperature (K) | 173(2) |
| Crystal system | Monoclinic |
| Space group | P21/n |
| Unit cell dimensions (Å, °) | |
| a | 9.1524(2) |
| b | 9.3816(2) |
| c | 18.8619(3) |
| β | 101.5050(10) |
| Volume (Å3), Z | 1587.02(5), 2 |
| Calculated density (g cm−3) | 2.25 |
| Absorption coefficient (mm−1) | 2.7 |
| F(000) | 1044 |
| Crystal size (mm3) | 0.27 × 0.23 × 0.11 |
| θ range for data collection (°) | 2.20–28.00 |
| h, k, l ranges | −12:12, −12:12, −24:24 |
| Reflections collected | 24192 |
| Independent reflections | 3833 |
| Rint | 0.0528 |
| Data / restraints / parameters | 3833 / 8 / 226 |
| Goodness-of-fit on F2 | 1.051 |
| Final R indices [I>2σ(I)] | R1 = 0.0284, wR2 = 0.0713 |
| R indices (all data) | R1 = 0.0341, wR2 = 0.0734 |
| Largest diff. peak and hole (e.Å−3) | 0.68 and −1.22 |
Table 2. Selected bond lengths (Å) and angles (°) for 1.
| Bond lengths (Å) | Angles (°) | ||
| Cd1−N1 | 2.358(3) | N1−Cd1−N6 | 177.20(9) |
| Cd1−N6 | 2.477(3) | N1−Cd1−Cl1 | 92.67(7) |
| Cd1−Cl1 | 2.5578(8) | N1−Cd1−Cl2 | 94.69(7) |
| Cd1−Cl2 | 2.5743(8) | N1−Cd1−Cl3 | 88.78(7) |
| Cd1−Cl3 | 2.6379(8) | Cl1−Cd1−Cl2 | 96.41(3) |
| Cd1−Cl4 | 2.6666(8) | Cl1−Cd1−Cl4 | 92.82(3) |
| Cd2−N2 | 2.277(3) | N2−Cd2−N2i | 180.0 |
| Cd2−Cl3 | 2.7182(7) | N2−Cd2−Cl3 | 84.61(7) |
| Cd2−Cl4 | 2.6724(8) | N2−Cd2−Cl4 | 89.17(8) |
Symmetry code: (i) – x + 1, − y + 1, − z + 1.
Results and Discussion
Reactions between L.HCl and ethanolic solution of cadmium(II) iodide provided colorless crystals of 1. This complex was characterized by IR, Raman, 1H NMR spectroscopy and X-ray crystallography. This complex is air-stable and soluble in DMSO and DMF.
Presence of the RNH2 group in 1 affect IR spectrum in two regions including 3317 and 3240 cm–1 for asymmetric and symmetric NH2 stretches and 1643 cm–1 for NH2 bending. The bands at 1697, 1281 and 1092 cm–1 which were assigned to the ν (C=N), ν (CN) and ν (NN), respectively,confirm the presence of the triazole ring.
Description of the crystal structure
The crystal structure of 1 was determined by X-ray single-crystal diffraction. The molecular graphic was drawn with ORTEP-III11.
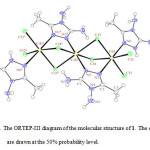 |
Figure 1: The ORTEP-III diagram of the molecular structure of 1. The ellipsoids are drawn at the 50% probability level. |
In the crystal structure of 1 (figure. 1), the three cadmium atoms have a distorted octahedral coordination environment. Four sites are occupied by chloride ions with the Cd–Cl bond lengths in the range of 2.5578(8)–2.7182(7) Å. The two other sites are occupied by two nitrogen atoms of L. The ligand L coordinated by two different coordination modes. Two chloride and a N–N bridges connect two adjacent cadmium atoms. The complex has a center of inversion on the copper atom and Ci symmetry.
Supplementary material
CCDC 913232 for [Cd3(L)2(HL)2(μ-Cl)4Cl4] contain the supplementary crystallographic data for this paper. These data can be obtained free of charge via www.ccdc.cam.ac.uk/conts/retrieving.html.
Acknowledgement
We are grateful to Payame Noor University of I. R. Iran for financial support.
References
- Patel N. B., Khan I. H. and Rajani S. D., Eur. J. Med. Chem., 45, 4293 (2010).
- Aromí G., Barrios L. A., Roubeau O. and Gamez P., Coord. Chem. Rev., 255, 485 (2011).
- Ghule V. D., Comput. Theor. Chem., 992, 92 (2012).
- Hakimi M., Mardani Z., Moeini K., Minoura M. and Raissi H., Z.Naturforsch., 66b, 1122 (2011).
- Hakimi M., Mardani Z., Moeini K., Mohr F., Schuh E. and Vahedi H., Z.Naturforsch., 67b, 452 (2012).
- Hakimi M., Mardani Z., Moeini K. and Fernandes M. A., J. Coord. Chem., 65, 2221 (2012).
- Yazdanbakhsh M., Hakimi M., Heravi M. M. and Boese R., Z. Anorg. Allg. Chem., 632, 2201 (2006).
- APEX-II, Bruker Advanced X-ray solutions (version 2.0-1), Bruker AXS Inc, Madison, Wisconsin (USA) 2005.
- SAINT-NT, Bruker Advanced X-ray solutions (version 6.0.) includes XPREP and SADABS, Bruker AXS Inc, Madison, Wisconsin (USA) 2005.
- SHELXTL, Bruker Advanced X-ray solutions (version 5.1.) includes XS, XL, XP, XSHELL, Bruker AXS Inc, Madison, Wisconsin (USA) 1999.
- Farrugia L. J., J. Appl. Cryst., 30, 565 (1997).

This work is licensed under a Creative Commons Attribution 4.0 International License.









