A Thermodynamic Study on the Binding of Human Serum Albumin with New synthesized Anticancer Pd(II) Complex
G. Rezaei Behbehani and L. Barzegar
Department of Chemistry, Imam Khomeini International University, Qazvin, Iran.
Article Received on :
Article Accepted on :
Article Published : 20 Oct 2016
The thermodynamics interaction between new synthesized anti-cancer drugcomplex, Pd2Py2, and HSA wasinvestigated at pH 7 by isothermal titration calorimetry. The extended solvation model was used to reproduce the enthalpies of HSA interaction withPd2Py2. The solvation parameters obtained from the new model was attributed to the structural change and anti-oxidant property of HSA. The binding parameters for the interaction of Pd2Py2 and HSA indicated that the considerable conformational changes in HSA were not observed after binding toPd2Py2.
KEYWORDS:Human serum albumin; Anti-cancer; Isothermal titration calorimetry; Binding parameters
Download this article as:| Copy the following to cite this article: G. Rezaei Behbehani G. R, Barzegar L. A Thermodynamic Study on the Binding of Human Serum Albumin with New synthesized Anticancer Pd(II) Complex. Orient J Chem 2012;28(4). |
| Copy the following to cite this URL: G. Rezaei Behbehani G. R, Barzegar L. A Thermodynamic Study on the Binding of Human Serum Albumin with New synthesized Anticancer Pd(II) Complex. Available from: http://www.orientjchem.org/?p=22679 |
Introduction
Human serum albumin (HSA) is the most abundant protein in the systemic circulation, with HSA comprising 60% in plasma. This protein can play a dominant role on the drug disposition and the efficiency. HSA has a high affinity to an extraordinarily diverse range of materials, such as drugs, metabolites, fatty acids and metal ions.1-4 HSA can bind and carry through the bloodstream many drugs, which are poorly soluble in water and it is responsible for the maintenance of blood pH, the drug disposition and efficacy, and the contribution of colloid osmotic blood pressure.4-6 The unique feature of albumin is its ability to bind a wide variety of compounds, mainly because of the availability of hydrophobic pockets inside the protein network and the flexibility of the albumins to adapt its shape.7-8 The crystallographic analysis of HSA revealed that this protein is a single-chain 66 kDa protein, which is largely α-helical, and consists of three structurally homologous domains that assemble to form a heart-shaped molecule.Each domain is a product of two subdomains, which are predominantly helical and extensively cross-linked by several disulfide bridges.9-11
The development of palladium anticancer drugs has not been promising and their design has mainly been based on the structure-activity relationship used for platinum anticancer drugs as well as good models for the analogous Pt(II) complexes in solution. Pd (II) complexes are expected to have lower kidney toxicity than cisplatin due to the inability of proteins in the kidney tubules to replace the tightly bound chelate ligands of Pd (II) with sulfydril groups.12-14 Studies have shown that Pd (II) complexes are very effective in inhibiting proliferation in several tumor cell lines.15-17
This work represents the most comprehensive study on the interactions between a new design anticancer drug of Pd2Py2 with carrier blood protein of HSA and provides new evidence for validity of the extended solvation model and more insights into the interactions of Pd2Py2 with HSA for further understanding of the effects of metal based drugs on the stability and the structural changes of carrier proteins.
Materials And Method
HSA was obtained from Sigma-Aldrich (Taiwan, China) and Pd2Py2was synthesized in our laboratory.17 Protein concentrations were determined from absorbance measurements at 277 nm in 1 cm quartz cuvettes. All other materials and reagents were of analytical grade, and solutions were made in 50 mM buffer phosphate using double-distilled water.
The isothermal titration calorimetric experiments were carried out on a VP-ITC ultra sensitive titration calorimeter (MicroCal, LLC, Northampton, MA). The microcalorimeter consists of a reference cell and a sample cell of 1.8mL in volume, with both cells insulated by an adiabatic shield. All solutions were thoroughly degassed before use by stirring under vacuum. The sample cell was loaded with HSA solution (60 M ) and the reference cell contained buffer solution. The solution inthe cell was stirred at 307 rpm by the syringe (equipped with micro propeller)filled withPd2Py2solution (1.2mM) to ensure rapid mixing. Injections werestarted after baseline stability had been achieved. The titration of HSA withPd2Py2 solution involved 30 consecutive injections of Pd2Py2 solution, thefirst injection was 5 μL and the remaining ones were 10 μL. In all cases, each injection wasdone in 6 s at 3-min intervals. To correct the thermal effects due to Pd2Py2 dilution, control experiments were done in which identical aliquotswere injected into the buffer solution with the exception of HSA. In the ITC experiments, the enthalpy changes associated with processes occurring at a constant temperature are measured. The measurements were performed at a constant temperature of 27.0±0.02 ◦C and the temperature was controlled using a Poly-Science water bath.
Result And Discussion
We have shown previously that the heart of the macromolecules + ligands interactions can be reproduced by Eq. 1in the aqueous solvent system.18-29
qis the heat of HSA + Pd2Py2interaction at certain ligand concentrations and qmax represents the heat value upon saturation of all HSA. The parameters
exhibit the HSA + Pd2Py2 stability in the low and high Pd2Py2 concentrations respectively. The positive values of
show that HSA is stabilized by Pd2Py2. can be expressed as follows
[Pd2Py2]is the concentration of ligand after every injection and [Pd2Py2]max is the maximum concentration of Pd2Py2 upon saturation of all HSA.The heats of HSA+Pd2Py2 interactions, q, were fitted to Eq.1 over the whole Pd2Py2compositions. In the fitting procedure, p was changed until the best agreement between the experimental and calculated data was approached. If the binding of ligand at one site increases the affinity for ligand at another site, the macromolecule exhibits positive cooperativity (p>1). Conversely, if the binding of ligand at one site lowers the affinity for ligand at another site, exhibits negative cooperativity (p<1). If the ligand binds at each site independently, the binding is non-cooperative (p=1).LA and LB are the unbound and bound ligand contributions to the heats of dilution in the absence of HSA. LA and LBcan be calculated from heats of dilution of Pd2Py2 in water, , as follows
The optimized
values are recovered from the coefficients of the second and third terms of Eq. 1. The small relative standard coefficient errors and the high r2values (0.99999) support the method (fig. 1). The binding parameters for HSA+Pd2Py2interactions recovered from Eq. 1 were listed in Table 2.
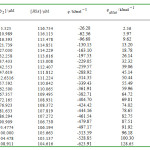 |
Table 1: heats of HSA+ Pd2Py2 interaction, q , at 300 K in 50mM phosphate buffer solution of pH=7, is the heat of dilution of with water. The precision is ± 0.1 µJ or better. |
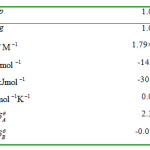 |
Table 2: binding parameters for HSA+ interaction recovered from Eq. 1at pH=7. The positive value of δθA show that stabilizes the HSA structure in the low concentration of Pd2Py2. Click here to View table |
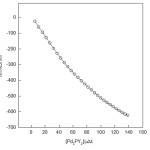 |
Figure 1: Comparison between the experimental heats, q, (O), for HSA+ interactions and calculated data (lines) via Eq. 1. is concentration of in μM at pH=7. |
For a set of identical and independent binding sites, it is possibleto use Eq. (5) for calculation of Kd and ”g” in a very simple way as follows
Where . q represents the heat value at a certain ligand and biomolecule concentration.M0 and L0 are total concentrations of HSA and ligand, respectively.qmax represents the heat value upon saturation of all HSAmolecule.Kd is the dissociation equilibrium constant for the equilibrium
![]()
If q and qmax are calculated per mole of biomacromolecule then the molar enthalpy of binding for each binding site ( ) will be:
![]()
The standard Gibbs free energy, ∆G°, can be calculated from association constant ![]()
![]()
Where is the appearance association equilibrium constant as a function of Pd- anti-cancer concentration. Therefore for the first time, we managed to calculate and values with using one set of experimental data in one temperature. All thermodynamic parameters of ligand binding to HSA are summarized in Table 2.
Conclusion
It has been confirmed that the extended coordination model, via equation 1 will satisfactorily reproduce the heats of HSA+Pd2Py2 interaction. Prediction of number of binding sites on HSA molecule, structural changes, determining the binding enthalpies and associated binding constants for such a complicated system accurately, make this theory the most powerful one. and values are positive and negative respectively (table 2), indicate that considerable conformational changes in HSA were observed due to the interaction with Pd2Py2.The negative value of shows that the HSA structure is destabilized in the high concentration of Pd2Py2. The negativevalue of molar enthalpy (-17889kJ mol-1 at 300 K) suggest that the binding process is only enthalpy-driven, indicating that electrostatic interaction is more important in the interaction.
Acknowledgements
Financial support from the Universities of Imam Khomeini (Qazvin) and Tehran are gratefully acknowledged.
References
- Cheng, F.Q.; Wang, Y. P.; Li, Z. P.; Dong, C.: Fluorescence study on the interaction of human serum albumin with bromsulphalein. Spectrochimica. Acta Part A: 65, 1144–1147 (2006)
- Cui, F.; Wang, J.; Cui, Y.; Li, J.; Fan, J.; Yao, X.: Binding of Human Serum Albumin to N-(p-Ethoxy-phenyl)-N′-(1-naphthyl) thiourea and Synchronous Fluorescence, Determination of Human Serum Albumin. Anal. Sci. 23, 719-725 (2007)
- Cui, F.L.; Fan, J.; Li, W.; Fan, Y.C.; Hu, Z.D.: Fluorescence spectroscopy studies on 5-aminosalicylic acid and zinc 5-aminosalylicylate interaction with human serum albumin. J. Pharm. Biomed. Anal. 34, 189–197 (2004)
- Das, P.; Mallick, A.; Haldar, B.; Chakrabarty, A.; Chattopadhyay, N.: Fluorescence resonance energy transfer from tryptophan in human serum albumin to a bioactive indoloquinolizine system. J. Chem. Sci. 119, 77–82 (2007)
- Rieutord, A.; Bourget, P.; Troche, G.; Zazzo, J. F.: In vitro study of the protein binding of fusidic acid: a contribution to the comprehension of its pharmacokinetic behaviour. Int. J. Pharm. 119, 57-61 (1995)
- Bogdan, M.; Pirnau, A.; Floare, C.; Bugeac, C.: Binding interaction of indomethacin with human serum albumin. J. Pharm. Biomed. Anal. 47, 981–984 (2008)
- Colmenarejo, G.: In silico prediction of drug-binding strengths to human serum albumin. Med. Res. Rev. 23, 275-301 (2003)
- Bordbar, A.K.; Sohrabi, N.; Gharibi, H.: Binding Set Analysis for Interaction of Human Serum Albumin with CethylTrimethylammonium Bromide. Bull. Korean Chem. Soc. 25, 791-795 (2004)
- Yue, Y.; Chen, X.; Qin, J.; Yao, X.: A study of the binding of C.I. Direct Yellow 9 to human serum albumin using optical spectroscopy and molecular modeling. Dye. Pigments 79, 176-182 (2008)
- Zhang, G.; Que, Q.; Pan, J.; Guo, J.: Study of the interaction between icariin and human serum albumin by fluorescence spectroscopy. J. Mol. Struct. 881, 132-138 (2008)
- Wang, T.; Xiang, B.; Wang, Y.; Chen, C.; Dong, Y.; Fang, H.; Wang, M.: Spectroscopic investigation on the binding of bioactive pyridazinone derivative to human serum albumin and molecular modeling. Coll. Surf. B: Biointerfaces 65, 113-119 (2008)
- Budzisz, E.; Krajewska, U.; Rozalski, M.: Cytotoxic and proapoptotic effects of new Pd (II) and Pt (II)-complexes with 3-ethanimidoyl-2-methoxy-2H-1, 2-benzoxaphosphinin-4-ol-2-oxide. Pol. J. Pharm.56, 473-478 (2004)
- Mansouri-Torshizi, H.; Srivastava, T.S.; Perekh, H.K.; Chitnis, M.P.: Synthesis, spectroscopic, cytotoxic and DNA-binding studies of binuclear Pt and Pd (II) complexes of meso- diaminoadipic and meso-diaminosuberic acids. J. Inorg. Biochem. 45, 135-48 (1992)
- Zhao, G.; Sun, H.; Lin, H.; Zhu, S.; Su, X.; Chen,Y.: Palladium(II) complexes with N,N′-Dialkyl-1,10-phenanthroline-2,9-dimathanamine: synthesis, characterization and cytotoxic activity. J. Inorg. Biochem. 72, 173-177 (1998)
- Divsalar, A.;Saboury, A.A.; Yousefi, R.; Moosavi-Movahedi, A.A.; Mansoori-Torshizi, H.: Spectroscopic and cytotoxic studies of the novel designed palladium(II)-complexes: β-Lactoglobulin and K562 as the targets. Int. J. Biol. Macro. 40, 381-86 (2006)
- Genova, P.; Varadinova, T.; Matesanz, A.I.; Matesanz, D.; Souza,P.: Toxic effects of bis(thiosemicarbazone) compounds and its palladium(II) complexes on herpes simplex virus growth. Toxic. Apply. Pharm.197, 107-112 (2004)
- Mansoori-Torshizi, H.; Islami-Moghaddam, M.; Saboury, A. A.: A microcalorimetry, spectroscopy study on the interaction of BSA with 2,2’-Bipyridine Octylglycinato palladium (II) nitrate. Acta. Biochim. Biophys. Sci. 35, 886-90 (2003)
- Rezaei Behbehani, G.: Application of a new method to reproduce the enthalpies of transfer of NaI from water to aqueous methanol, ethanol and iPrOH solvent systems at 289.15K. Bull. Korean Chem. Soc.2, 238-240 (2005)
- Rezaei Behbehani, G.: Application of the new solvation theory to reproduce the Enthalpies of transfer of LiBr, tetrabuthylammonium bromide and tetrapenthylamonium bromide from water to aqueous acetonitrile at 298 K.Acta Chim. Slov.52, 282-285 (2005)
- Rezaei Behbehani, G.; Tazikeh, E.; Saboury, A.A.: Using the New Developed Equation to Reproduce the Enthalpies of Transfer of Urea from Water to Aqueous Ethanol, Propan-1-ol and Acetonitrile at 298 K. Bull. Korean Chem. Soc.2, 208-210 (2006)
- Rezaei Behbehani, G.; Ghamamy, S.: Enthalpies of transfer of formamide, N-methylformamide and N,N-dimethylformamide from water to aqueous acetonirile mixtures at 298K.Thermochim. acta444, 71-76 (2006)
- Rezaei Behbehani, G.; Ghamamy, S.: Waghorne, W.E.: Enthalpies of transfer of acetonitrile from water to aqueous methanol, ethanol and dimethylsulphoxide mixtures at 298.15 K. Thermochim.acta,448, 37-42 (2006)
- Rezaei Behbehani, G.; Tazikeh, E.; Saboury, A.A.: Using the Extension Coordination Model (ECM) to reproduce the enthalpies of transfer of tetraethylurea from water to aqueous ethanol, propan-1-ol and acetonitrile at 298 K. Acta Chim. Slov.53, 363-369 (2006)
- Rezaei Behbehani, G.; Saboury, A. A.: Using a new solvation model for thermodynamic study on the interaction of nickel with human growth hormone.Thermochim. acta452, 76-79 (2007)
- Rezaei Behbehani, G.; Saboury, A. A.; Fallah baghery, A.: A Thermodynamic Study on the Binding of Calcium Ion with Myelin BasicProtein. J. Solution Chem. 36, 1311-1320(2007)
- Rezaei Behbehani, G.; Saboury, A.A.; Taleshi, E.: A Comparative Study of the Direct Calorimetric Determination of the Denaturation Enthalpy for Lysozyme in Sodium Dodecyl Sulfate and Dodecyltrimethylammonium Bromide Solutions. J. Solution Chem. 37, 619-629(2008)
- Rezaei Behbehani, G.; Saboury, A.A.: A thermodynamic study on the binding of Magnesium with Human Growth Hormone, Consideration of the new extended coordination model solvation parameters. J. Therm. Anal.Calorim. 89, 859-863 (2007)
- Rezaei Behbehani, G.; Saboury, A.A.; Takeshi, E.: Determination of partial unfolding enthalpy for lysozyme upon interaction with dodecyltrimethylammonium bromide using an extended solvation model. J. Mol. Recognit. 21, 132-135 (2008)
- Rezaei Behbehani, G.; Saboury, A.A.: A directcalorimetric determination of denaturation enthalpy for lysozyme in sodiumdodecylsulfate, J. Coll. Surf. B: Biointerfaces61, 224-228 (2007)
- Paal, K.; Muller, J.; Hegedus, L.: High affinity binding of paclitaxel to human serum albumin. Eur. J. biochem.268, 2187-2191 (2001)

This work is licensed under a Creative Commons Attribution 4.0 International License.












