Co-Ordination Ability of Vinylimidazole(Vim), Imidazole(Im), Indazole(In), Ammonia(NH3) and Dimethyl Sulphoxide (DMSO) Ligands in Ruthenium Complexes.
Rahul Sarma Baruah1, Rinki Moni Kalita2 and Chitrani Medhi2*
1Department of Applied Sciences, Chemical Science, Gauhati University.
2Department of Chemistry, Gauhati University Guwahati-781014, Assam, India.
Corresponding Author E-mail: chitranimedhi@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/360617
Article Received on : 06-10-2020
Article Accepted on :
Article Published : 30 Dec 2020
Few essential electronic properties of potential anti-cancer complexes are examined using quantum mechanical calculations. The co-ordination ability of these ligands with the central Ru metal has been assessed. The ionization energies and HOMO-LUMO gaps are used to explain the coordinate bond formation between ligand and Ru. The variation of charges from natural population analysis(NPA) of the donor sites of ligands is fairly similar to the trends in the coordinate bond distances despite the presence of other ligands in these Ru complexes. Slight variation of coordinate bonds (Ru-donor site) has been found. In addition, Cl dissociation energies from these complexes do not vary much and the trend of the energy values are similar to oxidation energies of these complexes. The NPA charges and HOMO-LUMO electron density mapping indicate that the electron density around Ru metal is increasing, which suggests that the migration of electron density is towards Ru.
KEYWORDS:Anticancer; Coordinate Bond; DNA Binding; Imidazole; NAMI; Ru Complexes; Vinyl Imidazole
Download this article as:| Copy the following to cite this article: Baruah R. S, Kalita R. M, Medhi C.Co-Ordination Ability of Vinylimidazole(Vim), Imidazole(Im), Indazole(In), Ammonia(NH3) and Dimethyl Sulphoxide (DMSO) Ligands in Ruthenium Complexes. Orient J Chem 2020;36(6). |
| Copy the following to cite this URL: Baruah R. S, Kalita R. M, Medhi C.Co-Ordination Ability of Vinylimidazole(Vim), Imidazole(Im), Indazole(In), Ammonia(NH3) and Dimethyl Sulphoxide (DMSO) Ligands in Ruthenium Complexes. Orient J Chem 2020;36(6). Available from: https://bit.ly/2JmdwyR |
Introduction
Certain molecules are commonly used in coordination chemistry, particularly in the synthesis of important Ruthenium complexes. The compounds with a five-memberred heterocyclic ring, i.e imidazole derivatives have been found as anti-fungal, anti-malarial and anti-microbial agents1-4. Therefore, several investigations aim to synthesize complexes with these ligands with an intention of getting effective anti-cancer agents. Buton subsequent evaluation of medicinal properties, non-drug complexes are identified, which indicate that concrete fundamental analysis is required for the synthesis metal complexes. The ligands,Vinyl Imidazole(VIm), Imidazole(Im), Indazole(In), ammonia(NH3) and Dimethyl sulphoxide (DMSO) have been used as coordinating ligands of some complexes 5-6. These ligands are found in some anticancer agents KP1019 [InH[RuCl4(In)2] [Indazolium trans- tetrachlorido bis (indazoleruthenate(III)], KP418[ImH[RuCl4(Im)2][Imidazolium trans- [tetrachloride bis(imidazole)ruthenate(III)], NAMI[Na[RuCl4(DMSO)Im][Sodium trans-tetrachlorido dimethyl suphoxide imidazole ruthenate(III), and [Ru(VIm)(DMSO)Cl4] [Tetrachlorido dimethylsulphoxide vinylimidazole ruthenate(III)].
Im and VIm are important ligands with two conjugate donor sites for coordination with Ru metal. Several molecules are identified to be important in medicine and in the synthesis of bio-active complexes with Ru metal7-10. It is essential to explore the details of their structures, coordination ability and electronic behaviour of molecules prior to the synthesis of Ru complexes for procuring specific use in cancer therapy. Many aspects of understanding the co-ordination ability of a molecule are essential for interpreting the bonding of these molecules towards certain metals such as their electronic properties. Following which, the stability of the complexes or the dissociation ability of ligand from the metal can be interpreted. Some N-donor aromatic ligands are known for efficient biological activity and are commonly used in metal-based drugs. In and Im derivatives are good antimicrobial, antifungal agents and these ligands are frequently usedin several metal complexes particularly in the synthesis of anti-cancer and anti-HIV drugs5-11. In addition, highly reactive ligands in abiological system are also used in the synthesis of potential drugs. The main factors for the efficiency of a drug can depend on certain physiochemical properties; (a) the complexes must be highly stable in the bio-system, so that it can overcome other complex reactions,and adverse effects on the biological property. Hence the stability of the metal complex is very important. So donor-acceptor behaviour of biologically important ligands towards metal must be clearly understood. (b) The electronic lability of ligands after complex formation with metal is generally different from the free ligand. In that case, comparison of electronic lability of ligands and the metal complex of ligands is necessary. will help in the understanding of metal coordination as well as ligand binding with bio-system. In addition, the electronic properties of ligands,such as metal coordination,contribute to the stability of metal complexes4,8-14. Therefore, few ligands found in some Ru-complexes are chosen for comparison in this study. Hence, the electronic behaviour of these complexes are analysed in the present study even if the electronic behaviour and drug-like propertymay not be linkeddirectly. The stability of the complexes having these ligands must be unique, since the drug efficiency is generally assessed without biasing inclusion of the decomposed ligand subunits and free cationic metal interactions with bio-system11-15. The unstable form of the complex may generate polar and cationic metal centre in solution, and it becomes difficult to reach the biological target when passing through the lipid bilayer asit stickson the cell membrane5,7,14-17. In this respect, the feature of one electron oxidation energy and Frontier orbital energies i.e HOMO and LUMO of the ligandsmay also be used to interpret certain electronic behaviour during coordination with Ru metal.
The structural framework of these ligands when coordinated to metal is also another important factor, and Ru complexes can form Imand VIm complexes in several proportions and also can form mixed ligand complexes with other ligands. So in this study, we have taken some ligands to explore the basic electronic properties while co-ordinating with Ru metal. The main goal of this study is to gain basic ideas how the metal ligand bonding is influenced by the electronic distribution around the donor site. As we know that the chemical properties of molecules depend on many parameters, i.e electronic and acid/base behaviours, which are usually two inseparable factors for undergoing chemical reactions. So, in this study we explore certain fundamental properties of important ligands found in potential anti-cancer Ru complexes.
It should be noted that chlorine dissociation and Ru coordination with the nearest donor site of bio-system are simultaneous processes. Indeed this area has been highlighted in the study of some complexes, but the stability of Rucomplexes for generating Cl before binding with the target is not well assessed. So some ruthenium complexes have been taken for examining this reaction pathway. Resolving well defined pathway of these complexes for understanding the coordination ability of Ruthenium with the binding site may probably relevant to predicting susceptibility towardstarget bio-system.
In this study KP1019 [InH[RuCl4(In)2] [Indazolium trans- tetrachlorido bis (indazoleruthenate(III)], KP418[ImH[RuCl4(Im)2] [Imidazolium trans- [tetrachloride bis(imidazole)ruthenate(III)], Na[RuCl4(DMSO)Im][Sodium trans-tetrachlorido dimethyl suphoxide imidazole ruthenate(III)], and [Ru(VIm)(DMSO)Cl4][Tetrachlorido dimethylsulphoxide vinylimidazole ruthenate(III)] are explored and the dissociation ability of Cl from these complexes is further examined.
Computational Details
The structures of the ligands are shown in Figs.1(a-e). The structures consist of lone pair electron donor sites in heterocyclic ring and additional π-bond in VIm. The coordination ability of two N-sites (N2 and N4) of Im andVImmay be considered for stabilizing the Ru-complexes with these ligands. Similarly, distinct electron donor sites are present in the common ligands such as In, ammonia and DMSO. Complete geometry optimization of ligands and Ruthenium complexes are performed with B3LYP/SDD route using Gaussian programme code(09)19. Natural population analysis (NPA) has been carried out for these ligands and complexes.
The one electron ionization energy may be taken as a simple technique to examine the coordination ability of the ligands(L). Further studies on the 3D electron density mapping are also carried out to analyse the position of HOMO and LUMO in the ligands. The ionization energies (IE) of the ligandsare calculated from the energies of ligands (EL ) and energies of ionised ligand(EL+ ) as shown in the following equation.
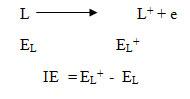
There are quite a few electron donor sites in some ligands. So electron donation ability and the NPA charges at these sites maybe used to indirectly link the coordination ability towards the metal. The decrease of electron density in L+ compared to L of the donor sites will indicate most effective site as well as the nature of electron density migration among various donor sites. Hence, natural population analysis has been carried out for these ligands. Alternately frontier orbital 3D mapping may be useful for explaining the electronic features around these ligands. All the calculations were carried out with Gaussian 09 programme code and Gauss View 4.118.
Dissociation energies of chlorine are calculated from the following mechanisms.
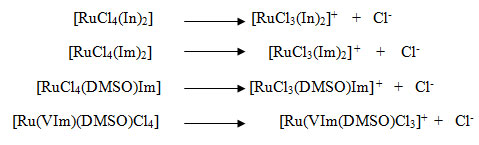
Four complexes, (1)KP1019: InH[RuCl4(In)2] (Indazolium trans-tetrachloridobis(indazole) ruthenate(III)]) (2) KP418: ImH[RuCl4(Im)2] (imidazolium trans-[tetrachloridobis (imidazole) ruthenate(III)]) (3) Nami: Na[RuCl4(DMSO)Im] (sodium tetra chloride dimethyl suphoxide imidazole ruthenate(III)) (4) Ru(VIm)(DMSO)Cl4] (Tetra chloride dimethyl sulphoxide vinyl imidazole ruthenate(III)) are taken in this study.
Results and Discussion
The ligands, VIm, Im, In, NH3 and DMSO are chosen to compare the coordination ability with Ru (Figs. 1(a)-(e)). The computed NPA values using B3LYP/SDD route in the Gaussian 09 are reported in Tables 1-5, and the variation of charges at the donor atom of the ligand is clearly shown. Again,for the purpose of comparison of the NPA charges of these ligands, the NPA charges of ionised ligands are also calculated and it will show the extent of charge migration on the atoms of ligands particularly at the donor atoms. The donor atoms that involve in coordination with Ru are found significantly different. It indirectly shows the lability of electron from the donor site, i.e large variation at N atomsare found for VIm (N2 and N4) and Im (N1 and N4) are observed. The maximum values of charges are observed at these Natomic sites of these two ligands,which in principle are the favourable coordination sites with Ru. Furthermore, for other ligands( In, Ammonia and DMSO), the NPA charges are given in Tables 3-5, the donor atomic sites are found well defined charge accumulation compared to the other atoms, hence the coordination bond formation with Ru can be understood from the negative values of charges on donor sites. The analysis of donor atomic sites based on NPA values for bonding with Ru is not be always possible if steric factors are involved during complex formation. From the orientation of donor sites particularly for Im and VIm, any observable steric hindrance is not found and the most favourable N donor atomic site that can directly involve in coordination with Ru, which can be identified from the NPA charge (Tables 1 and 2). The NPA charges of these two sites are not drastically different but more negative charge on coordinating N atom with Ru is found (Figs. 2(a, b)). As such, NPA charges may indirectly related to the bonding ability of these two ligands.Similar analysis have been performed for the commonly found ligands in potential anticancer agents, i.e In, DMSO and NH3, where donor sites are found coordinated in some Ru complexes. It demonstrates that the NPA charges are useful for identifying the preferable site for coordination with Ru with the exception for DMSO, where O and S can form coordinate bonds with Ru (Tables 1-5)12.
 |
Figure 1: Structures of the ligands, (a) Vinyl Imidazole(VIm), (b) Imidazole(Im), (c) Ammonia(NH3), (d) Indazole(In) and(e) DMSO. |
Table 1: Computed NPA charges on the atoms of VIm and VIm+.
|
Atoms |
Charges on VIm |
VIm+ |
|
C1 |
-0.061 |
0.353 |
|
N2 |
-0.513 |
-0.346 |
|
C3 |
-0.064 |
0.140 |
|
N4 |
-0.449 |
-0.187 |
|
C5 |
0.223 |
0.443 |
|
C6 |
0.009 |
0.017 |
|
C7 |
-0.434 |
-0.165 |
|
H8 |
0.219 |
0.132 |
|
H9 |
0.219 |
0.137 |
|
H10 |
0.208 |
0.124 |
|
H11 |
0.216 |
0.147 |
|
H12 |
0.222 |
0.118 |
|
H13 |
0.206 |
0.088 |
Table 2: Computed NPA charges on the atoms of Im and Im+.
|
Atoms |
Charges on Im Im+ |
|
|
N1 |
-0.528 |
-0.236 |
|
C2 |
-0.072 |
0.184 |
|
C3 |
-0.077 |
0.325 |
|
N4 |
-0.607 |
-0.306 |
|
C5 |
0.210 |
0.411 |
|
H6 |
0.217 |
0.130 |
|
H7 |
0.218 |
0.132 |
|
H8 |
0.207 |
0.129 |
|
H9 |
0.432 |
0.231 |
Table 3: Computed NPA charges on the atoms of DMSO and DMSO+ .
|
Atoms |
Charges on DMSO |
DMSO+ |
|
C1 |
-0.732 |
-0.458 |
|
S2 |
0.895 |
1.235 |
|
C3 |
-0.732 |
-0.425 |
|
H4 |
0.227 |
0.147 |
|
H5 |
0.238 |
0.161 |
|
H6 |
0.247 |
0.162 |
|
O7 |
-0.855 |
-0.251 |
|
H8 |
0.227 |
0.136 |
|
H9 |
0.247 |
0.147 |
|
H10 |
0.238 |
0.147 |
Table 4: Computed NPA charges on the atoms of NH3 and NH3+.
|
Atoms |
Charges on NH3 |
NH3+ |
|
N1 |
-1.182 |
0.334 |
|
H2 |
0.394 |
0.189 |
|
H3 |
0.394 |
0.236 |
|
H4 |
0.394 |
0.240 |
Table 5: Computed NPA charges on the atoms of In and In+.
|
Atoms |
Charges on In |
In+ |
|
C1 |
-0.121 |
-0.064 |
|
C2 |
-0.026 |
-0.051 |
|
C3 |
-0.327 |
-0.169 |
|
C4 |
-0.102 |
-0.031 |
|
C5 |
0.330 |
0.132 |
|
C6 |
-0.059 |
-0.175 |
|
C7 |
-0.414 |
-0.187 |
|
N8 |
-0.483 |
-0.244 |
|
N9 |
-0.094 |
0.945 |
|
H10 |
0.228 |
0.158 |
|
H11 |
0.168 |
0.120 |
|
H12 |
0.133 |
0.093 |
|
H13 |
0.170 |
0.111 |
|
H14 |
0.209 |
0.128 |
|
H15 |
0.386 |
0.234 |
 |
Figure 2: Structures of Complexes, (a) KP1019 (b) KP418 |
The complexes of these ligands with Ru are shown in Figs 2(a-d), and coordination distances of the optimized geometries are shown in Tables 7-10.Comparison of net charges of donor sites of several ligands can be useful to predict the most preferred site as well as the extent of electron donation towards Ru metal. Hence, the variation of NPA charges of only the donor atomic sites of these ligands can be differentiated from the values shown in Table 11.
Table 7: Computed bond lengths(Ru-X, X=Ru coordinated atoms) and NPA charges on coordinated atoms with ruthenium(III) in KP1019(InH[RuCl4(In)2]).
|
Complexes |
Dissociated complexes |
||||||
|
Bond lengths (Ao) |
NPA charges |
Bond lengths (Ao) |
NPA charges |
||||
|
Atoms
|
Charges |
Atoms |
Charges |
||||
|
Ru – Cl20 |
2.44 |
Cl20 |
-0.05 |
Ru – Cl20 |
2.42 |
Cl20 |
0.04 |
|
Ru – Cl21 |
2.44 |
Cl21 |
0.15 |
Ru – Cl21 |
2.42 |
Cl21 |
0.12 |
|
Ru – Cl22 |
2.44 |
Cl22 |
0.02 |
Ru – Cl22 |
2.42 |
Cl22 |
0.042 |
|
Ru – Cl23 |
2.44 |
Cl23 |
0.15 |
Ru – Cl23 Dissociated |
Cl23 Dissociated |
|
|
|
Ru – N13 |
2.02 |
N1 |
-0.10 |
Ru – N13 |
2.00 |
N13 |
-0.30 |
|
Ru – N11 |
2.02 |
N11 |
-0.12 |
Ru – N11 |
2.00 |
N11 |
-0.26 |
|
Ru |
0.43 |
Ru |
0.29 |
||||
Table 8: Computed bond lengths(Ru-X, X=Ru coordinated atoms) and NPA charges on coordinated atoms with ruthenium(III) in KP418(ImH[RuCl4(Im)2]).
|
Normal complexes
|
Cl Dissociated complex |
|||||||
|
Bond lengths (Ao) |
NPA charges |
Bond lengths (Ao) |
NPA charges |
|||||
|
Atoms |
Charges |
|
Atoms |
Charges |
||||
|
Ru – Cl12 2.43 |
Cl12 |
0.17 |
Ru – Cl12 |
2.42 |
Cl12 |
0.19 |
||
|
Ru – Cl13 2.43 |
Cl13 |
0.17 |
Ru – Cl13 |
2.42 |
Cl13 |
0.21 |
||
|
Ru – Cl14 2.43 |
Cl14 |
0.17 |
Ru – Cl14 |
2.42 |
Cl14 |
0.29 |
||
|
Ru – Cl15 2.43 |
Cl15 |
0.15 |
Ru – Cl15 Dissociated |
Cl15 |
Dissociated |
|||
|
Ru – N2 2.03 |
N2 |
-0.25 |
Ru – N2 |
2.00 |
N2 |
-0.47 |
||
|
Ru – N5 2.03 |
N5 |
-0.22 |
Ru – N5 |
2.00 |
N5 |
-0.47 |
||
|
|
Ru |
0.100 |
Ru |
0.18 |
||||
Table 9: Computed bond lengths(Ru-X, X=Ru coordinated atoms) and NPA charges on coordinated atoms with Ruthenium(III) in NAMI(Na[RuCl4(DMSO)Im] ) .
|
Complex |
Cl Dissociated complex |
||||||
|
Bond lengths (Ao) |
NPA charges Atoms charges |
Bond lengths (Ao) |
NPA charges Atoms charges |
||||
|
Ru – Cl2 |
2.43 |
Cl2 |
0.26 |
Ru – Cl2 |
2.43 |
Cl2 |
0.25 |
|
Ru – Cl3 |
2.43 |
Cl3 |
0.41 |
Ru – Cl3 |
2.43 |
Cl3 |
0.25 |
|
Ru – Cl12 |
2.43 |
Cl12 |
0.01 |
Ru – Cl12 |
2.44 |
Cl12 |
0.26 |
|
Ru – Cl13 |
2.44 |
Cl13 |
0.17 |
Ru – Cl13 Dissociated |
Cl13 Dissociated |
|
|
|
Ru – S4 |
2.17 |
S4 |
0.86 |
Ru – S4 |
1.17 |
S4 |
1.71 |
|
Ru – N5 |
2.03 |
N5 |
-0.20 |
Ru – N5 |
2.03 |
N5 |
– 0.47 |
|
Ru |
0.07 |
Ru |
-0.08 |
||||
Table 10: Computed bond lengths(Ru-X, X=Ru coordinated atoms) and NPA charges on coordinated atoms with Ruthenium(III) in [Ru(VIm)(DMSO)Cl4].
|
Complex Bond lengths (Ao)NPA charges Atoms Charges
|
Cl dissociated complex NPA charges Bond lengths (Ao) Atoms Charges |
|||||
|
Ru – Cl2 |
2.03 |
Cl2 |
-0.68 |
Ru – Cl2 2.04 |
Cl2 |
1.34 |
|
Ru – Cl3 |
2.12 |
Cl3 |
0.32 |
Ru – Cl3 2.09 |
Cl3 |
0.34 |
|
Ru – Cl14 |
2.19 |
Cl14 |
0.14 |
Ru – Cl14 2.03 |
Cl14 |
0.79 |
|
Ru – Cl15 |
2.15 |
Cl15 |
0.34 |
Ru – Cl13 Dissociated |
Cl15 Dissociated |
|
|
Ru – N4 |
1.96 |
N4 |
-0.08 |
Ru – N4 2.06 |
N4 |
-0.34 |
|
Ru – S5 |
2.02 |
S5 |
0.51 |
Ru – S5 2.02 |
S5 |
1.29 |
|
Ru |
-0.39 |
Ru |
-0.97 |
|||
Table 11: Comparison of the NPA charges on bonded atoms for different ligands and the oxidised ligands
|
Ligands
|
NPA charge(Free) |
NPA charge (Oxidised) |
||
|
1.Vinyl Imidazole(VIm) |
N2 N4 |
-0.513 -0.449 |
N2 N4 |
-0.346 -0.187
|
|
2.Imidazole(Im) |
N1 N4 |
-0.528 -0.607 |
N1 N4 |
-0.236 -0.306
|
|
3.Indazole(In) |
N8 N9 |
-0.483 -0.094 |
N8 N9 |
-0.244 -0.945
|
|
4.Dimethylsulphoxide(DMSO) |
S2 O7 |
0.895 -0.854 |
S2 O7 |
1.235 -0.251
|
|
5.Ammonia |
N1 |
-1.182 |
N1 |
0.334 |
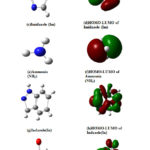 |
Figure 3: Structures of ligands and corresponding HOMO-LUMO electron densities; (a) Vinyl Imidazole(VIm)(b) HOMO-LUMO of Vinyl Imidazole(VIm)(c) Imidazole (Im)(d) HOMO-LUMO of Imidazole (Im) (e) Ammonia(NH3)(f) HOMO-LUMO of Ammonia(NH3) (g) Indazole(In)(h) HOMO-LUMO of Indazole(In)(i) DMSO(j) HOMO-LUMO of DMSO |
The electronic mobility of these ligands towards Ru can be related to the ionization energies [IE] of these ligands without specifically considering donor sites. Againthe frontier orbital electron density mapping and the maximum value of HOMO-LUMO gap can be used another important parameter for understanding the extent of donor-acceptor interaction in the Ru complexes (Table 6).
Table 6: Ionization energies(IE) and HOMO-LUMO gaps of ligands
|
Sl. No. |
Ligands |
IE (kcalmol-1) |
HOMO-LUMO gaps(kcalmol-1) |
|
1 |
Vinyl Imidazole(VIm) |
197.16 |
127.03 |
|
2 |
Imidazole(Im) |
204.57 |
157.09 |
|
3 |
Ammonia(NH3) |
225.63 |
396.28 |
|
4 |
Indazole(In) |
104.35 |
24.06 |
|
6 |
DMSO |
192.14 |
95.49 |
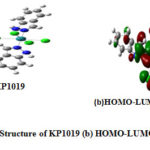 |
Figure 4: (a) Structure of KP1019 (b) HOMO-LUMO of KP1019 |
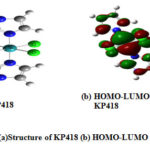 |
Figure 5: (a)Structure of KP418 (b) HOMO-LUMO of KP418 Click here to View figure |
From these values, it ispossible to identify the N donor site involved in forming coordinate bond with VIm, Im, In and NH3 (Figs. 2(a-d)). Table 6 reveals that the HOMO-LUMO gap of Im is smaller than VIm and can form Ru-N coordination bond easily. It indicates that HOMO-LUMO gaps as well as ionization energies of these ligands are, in fact, useful for analyzing the coordination bonding and stability of these complexes. The observation is different for DMSO where the NPA charges of O are negative and large positive NPA charge is found on S that may be taken for illustrating S as well as O coordination inthe synthesis of Ru complexes. Probably, Ru metal is a soft metal that may be more susceptible for S, which is easily polarization for forming coordinate bond. So, additional information may be collected from the HOMO-LUMO 3D diagram of the complexes of DMSO i.e in the complexes NAMI(Na[RuCl4(DMSO)Im]) and Ru(VIm)(DMSO)Cl4] where the electron density around DMSO, particularly S=O region can be taken for analysis. Although NPA charge of O is found negative due shifting of electron density from S to O, but large distribution of electron density within around S-O region of DMSO is clearly shown (Figs. 6(a,b) and 7(a,b)). It could be the reason why both S and O coordinated DMSO complexes of Ru are formed. Also there may be steric hindrance from the two methyl groups for coordinating with S under certain situation. Again, the coordination of Ru with any donor sites of nucleobase is expected during binding, but platinum metal of cisplatin coordinates with N7 of guanine during DNA binding. In such a situation, dissociation of Cl from the complex and subsequent binding with N7 of guanine is the competitive processes during drug action. Likewise, dissociation of Cl from these complexes is an essential step for Ru binding with the receptor, and such a reaction also could be dependent on the Ru coordination ability of these complexes. To this end, we have investigated the dissociation energies (DE) of Cl from these complexes and the values are shown in Table 6. Overall, it has been observed that the values of DE are not very different for these complexes. One can speculate that DE values are somewhat related to the oxidation energies of these complexes because the redox energies are not specifically for Ru center and these are the straightforward one electron transfer energies of the complex.So the dissociation of Cl from these Ru complexes is investigated. This is considered as another issue for development of DNA binding Ru complexes.
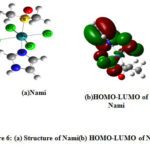 |
Figure 6: (a) Structure of Nami(b) HOMO-LUMO of Nami |
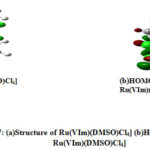 |
Figure 7: (a)Structure of Ru(VIm)(DMSO)Cl4] (b)HOMO-LUMO of Ru(VIm)(DMSO)Cl4] |
It can be seen that Ru-N coordination distances of ligands (except DMSO) vary significantly and in fact, shorter interaction distance indicates favourable bonding. It can be pointed out that NPA charges as well as the IE of ligands are somewhat important for explaining such coordination bonds. The observed differences of the coordination bond lengths may be indirectly used to compare the coordination ability of the ligands with Ru for these complexes. There will be stereoelectronic control in the formation of coordination bond, but NPA charges have been shown to determine fairly the formation of Ru-N bonds(Table 12).
Table 12: Computed redox energies different Ru complexes, dissociation energies of Cl and HOMO-LUMO gaps.
|
Complex |
Oxidation states |
Redox Energies (kcal/mol) |
DE (kcal/mol) |
HOMO-LUMO gaps(kcalmol-1)
|
|
1. KP1019 |
Ru(III)/ Ru(IV) |
447.780 |
400.22 |
167.98 |
|
|
Ru(III)/ Ru(II) |
-315.632 |
|
|
|
2.KP418 |
Ru(III)/ Ru(IV) |
521.452 |
404.17 |
166.10 |
|
|
Ru(III)/ Ru(II) |
-354.537 |
|
|
|
3..NAMI |
Ru(III)/ Ru(IV) |
525.217 |
400.22 |
95.63 |
|
|
Ru(III)/ Ru(II) |
-377.875 |
|
|
|
4. [Ru(VIm)(DMSO)Cl4] |
Ru(III)/ Ru(IV) |
431.538 |
421.24 |
52.71 |
|
|
Ru(III)/ Ru(II) |
-308.629 |
|
|
The coordination of Ru with these donor sites can be indirectly verified from Ru-N bond lengths. It may be due to electron lability of ligands, which there by contributes to the formation of coordination bond with Ru. The coordination of O in DMSO is possible when the charge densities of Ru increases on coordination with other donor ligands, then O will be active center for interaction Ru. Hence, coordination with Ru in some Ru complexes with DMSO ligands are O bonded with Ru. It is worth mentioning that the oxidation and reduction energies of the complexes are not equal, even if both the processes are opposite to each other. From the energy values, reduction may be a more feasible pathway than oxidation. However, the oxidation pathway is possible if a strong cationic group is present in the receptor or the binding site. Comparison of DE of Cl with that of redox energies has been done and we observed fairly similar variation ofenergy values. Moreover, oxidation energies are of similar trends with that of DE of Cl, and are almost equal for certain complexes. So choosing the ligand fragments for coordination with Ru is important to form stable Ru complexes. In this study, few ligandsare explored to examine the trends of IE, NPA charges and HOMO-LUMO. We therefore could extract useful information from this study i.e coordination capability of ligands with Ru, which may be useful for demonstrating the stability of Ru complexes.
Conclusion
The studies on VIm, Im,In, NH3 and DMSO have been taken to assess the coordination ability of these two ligands. The variation of oxidation energies shows similar trend with that of HOMO-LUMO gaps and IE. But the DE values of Cl from these complexes are almost the same. In addition, the donor ability of these ligands is indirectly assessed from the coordination distances of these donor sites towards Ru. The closer distance observed in VIm complexes predicts better coordination with Ru. This observation is relevant to monitor the stability of Ru complexes and effect of ligands on the redox energies of complexes.
Acknowledgement
The authors thank Department of Applied Sciences the Gauhati University for the computational work. There are no ethical and conflict issues in this work.
References
- Hartinger, C. G.; Zorbas-Seifried, S.; Jakupec, M. A.; Kynast, B.; Zorbas, H.; Keppler, B. K.; J. Inorg.Biochem.,2006,100,891.
CrossRef - (a) Kapitza, S., Pongratz, M., Jakupec, M. A.; Heffeter P.; Berger, W.; Lackinger, L.; Keppler, B. K.; Marian B. J.; Cancer Res. Clin. Oncol.,2005, 131,101.
CrossRef
(b) Nováková, O.; Kaspárková, J., Vrána, O.; van Vliet, P.M.; Reedijk, J.; Brabec,V.; Biochemistry,1995, 34, 12369.
CrossRef
(c) van Vliet, P. M., Haasnoot, J. G., Reedijk, J. Inorg.Chem., 1994, 33,1934. - Polec-Pawlak, K.; Abramski, J. K.; Semenova, O.; Hartinger, C. G.; Timerba ev , A R.; Keppler, B. K.; Jarosz M.; Electrophoresis, 2006,27,1128.
CrossRef - (a) Kostova I.; Ruthenium complexes as anticancer agents, Curr Med Chem.,2006, 13, 1085-1107.
CrossRef
(b) Bonin, A. M.; Yanez, J. A.; Fukuda, C.; Teng, X. W.; Dillon, C. T.; Hambley, T. W.; Lay, P. A.; Davies, N. M.; Cancer Chemother. Pharmacol.2010, 66,755−764.
CrossRef
(c) Psomas, G.; Kessissoglou, D. P.; Dalton Trans.2013, 42,6252−6276.
CrossRef
(d) Bergamo, A.; Masi, A.; Dyson, P. J.; Sava, G.; Int J. Oncol.2008,33,1281−1289.
(e) Hoffman K.; Imidazole and its Derivatives. Interscience Publisher,USA; 1953, 143- 145. - Thore S.N. ; and Gupta A. K.;Indian J Heterocycl Chem, 2010, 19,373.
- Eicher T., Hauptmann S., The Chemistry of Heterocycles: Structure, Reactions, Synthesis, and Applications. 2nd ed.; Wiley-VCH;2003.
CrossRef - Mirzaei J.; Amini M.; Pirelahi H.; and Shafiee A.; J Heterocycl Chem.,2008,45, 921.
CrossRef - Leijen, S.; Burgers, S. A.; Baas, P.; Pluim D.; Tibben, M.; vanWerkhoven, E., Alessio, E., Sava, G.; Beijnen, J. H.; Schellens, J. H. M.; Invest. New Drugs,2015, 33, 201−214.
CrossRef - Dillon, C. T.; Hambley, T. W.; Kennedy, B. J.; Lay, P. A.; Zhou, Q.; Davies, N. M.; Biffin, J. R.; Regtop, H. L.;Chem. Res. Toxicol. 2003, 16,28−37.
CrossRef - Dimiza, F., Papadopoulos, A. N., Tangoulis, V., Psycharis, V., Raptopoulou, C. P., Kessissoglou, D. P., Psomas, G. Dalton Trans.2010, 39,4517−4528.
CrossRef - James B. R., Ochiai E., Rempel G.I., Inorganic and Nuclear Chemistry Letters. 1971, 7 ,8 , 781–784.
CrossRef - Bratsos I.; Serli, B.; Zangranko E.; Katsaros, N.; Alessio, E.; Inorg. Chem.2007, 46 (3): 975–992.
CrossRef - Enzo A.; Chem. Rev.2004, 104 (9):4203–4242.
CrossRef - EnzoA.; Bentham Science Publisher; Giovanni Mestroni, Bentham Science Publisher; Alberta Bergamo, Current Topics in Medicinal Chemistry. 2004 ,15,1525–1535.
- ThangaduraiA.; MinuM.; WakodeS.; AgrawalS. and NarasimhanB. ,Medicinal Chemistry Research, 2012, 21, 1509–1523.
CrossRef - Salamone J.C.;Israel S.C., Taylo P.; Snider B, Polymer, 1973,14, 12, 639–644.
CrossRef - Cerecetto H.; Gerpe A., Gozalez M.; Aran V.J.; deOcariz C.O.;Mini Rev Med Chem2005, 5, 869–878.
CrossRef - M. J. Frisch.; G. W. Trucks.; H. B. Schlegel.; G. E. Scuseria.; M. A. Robb.; J. R. Cheeseman.; J. A. Montgomery, Jr.; T.Vreven.; K. N. Kudin.; J. C. Burant.; J. M. Millam, S.; S. Iyengar, J. Tomasi, V. Barone, B.; Mennucci, M.; Cossi, G.; Scalmani, N. Rega, G. A. Petersson, H.; Nakatsuji, M. Hada, M.;Gaussian, Inc., Pittsburgh PA, Gaussian 09, B.04 2009
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









