Validation and Optimization of Polyvinyl Alcohol-Functionalized Gold Nanoparticles for Producing Lactose-Free Dairy Products
1Department of Community Medicine and Pilgrims Health Care, Umm Alqura University, Makkah, Saudi Arabia
2Department of Biochemistry, School of Medicine, Batterjee Medical College for Sciences and Technology, Jeddah, Saudi Arabia.
Corresponding Author E-mail: shakeel.ansari@bmc.edu.sa
DOI : http://dx.doi.org/10.13005/ojc/370317
Article Received on : 22-Apr-2021
Article Accepted on :
Article Published : 26 Jun 2021
The present study demonstrates the synthesis of lactose-free dairy items by Kluyveromyces lactis β-galactosidase bound to polyvinyl alcohol (PVA)-modified gold nanoparticles (AuNPs). The size of AuNPs was analyzed by dynamic light scattering experiment. The developed AuNPs served as a stable matrix for enzyme immobilization which was observed by obtaining 88% immobilization yield. Km and Vmax were determined for soluble and immobilized enzyme by incubating them with varying concentrations of substrate. Our findings demonstrated that immobilization leads to an increase of Km and a decline in Vmax values for the enzyme attached to PVA-functionalized AuNPs. Moreover, the enzyme conjugated to surface functionalized AuNPs displayed exceptional conversion of lactose hydrolysis in batch reactors at 40 oC in contrast to its hydrolysis at 50 oC. Hence, the developed nanosystem [β-galactosidase-(PVA-modified AuNPs)] serves as an excellent model for suggesting its application in other biomedical applications, particularly for constructing lactose based biosensors.
KEYWORDS:β-galactosidase; Gold Nanoparticles; Lactose Hydrolysis; Modification; Polyvinyl Alcohol
Download this article as:| Copy the following to cite this article: Alshanberi A. M, Ansari S. A. Validation and Optimization of Polyvinyl Alcohol-Functionalized Gold Nanoparticles for Producing Lactose-Free Dairy Products. Orient J Chem 2021;37(3). |
| Copy the following to cite this URL: Alshanberi A. M, Ansari S. A. Validation and Optimization of Polyvinyl Alcohol-Functionalized Gold Nanoparticles for Producing Lactose-Free Dairy Products. Orient J Chem 2021;37(3). Available from: https://bit.ly/2UGtNE3 |
Introduction
Lactose is converted into its basic components, i.e., galactose and glucose, by the enzyme β-galactosidase, which is exclusively present in animals, microbes and plants. It is extensively used in biotechnology sectors and in constructing lactose-based biosensors.1,2 Furthermore, Kluyveromyces lactis β-galactosidase is homodimeric. It exhibited 45% glycosylation, and demonstrated temperature and pH-optima at 37 oC and pH 7.0, respectively.3 This enzyme demonstrates its exceptional utility in manufacturing lactose-free dairy products.4-7
Recently, the immobilization of enzymes on nanoparticles has attracted researchers as they impart large surface areas to bind greater quantity of enzyme on the nanosupports, hinder in unfolding the enzyme and promotes improvement in enzyme flexibility. It also balances the main elements for determining the efficacy of biocatalysts like effective enzyme loading, mass transfer resistance and surface area.8-12 Other notable advantages that are associated by using nanoparticles for enzyme immobilization are their easy separation from reaction mixture, catalyst recycling, continuous operations, increased stability and surface modification.13,14
Recently, gold nanoparticles were utilized by researchers to stabilize various biotechnologically and biomedically important enzymes.15-17 Apart from above-mentioned advantages, AuNPs also offered a great adsorption capacity, excellent thermal and mechanical stability. Additionally, they demonstrated rapid electrode kinetics, enhanced electronic properties and excellent biocompatibility to catalyze the biochemical process.18,19
Therefore, this study illustrates the modification of gold nanoparticles by polyvinyl alcohol (PVA) to immobilize β-galactosidase from Kluveromyces lactis. The particle dimension of the synthesized AuNPs was measured by dynamic light scattering technique. Free and conjugated β-galactosidases were characterized for Michaelis constant (Km) and maximum rate of reaction (Vmax). Moreover, the utility of β-galactosidase conjugated to PVA modified AuNPs was evaluated by lactose hydrolysis in batch reactors at 40 and 50 °C.
Experimental
Materials
Various buffers and β-galactosidase from K. lactis were procured from Sigma-Aldrich (USA). The substrate, o-nitrophenyl-β-D-galactopyranoside (ONPG), and poly vinyl alcohol (Mw 89,000-98,000, 99% hydrolyzed) was obtained from Sisco Research Laboratories, India. Double distilled water was used throughout the experimental methodology.
Characterization of AuNPs
Gold nanoparticles were prepared according to our recently published study.20 Additionally, dynamic light scattering (Malvern Zetasizer Nano ZS) technique was utilized to observe the particle size of the synthesized AuNPs.
Enzyme immobilization and determination of kinetic parameters
The initial step in enzyme immobilization involved the overnight incubation of β-galactosidase (2000 U) with the modified AuNPs at 30 °C under slow stirring conditions. The resulting conjugated enzyme was washed thrice with the assay buffer (100 mM, pH 7.0 potassium phosphate buffer) to detach the loosely bound enzyme. The developed nanosystem [β-galactosidase-(PVA-modified AuNPs)] was refrigerated at 4 °C in the assay buffer to proceed with the experiments. The following equation was used to calculate the enzyme immobilization yield:
% immobilization = B/A x 100 …………… (1)
where A denotes the enzyme units that were obtained by theoretical calculation [Loaded enzyme (X)-enzyme lost in washes (Y)] to achieve immobilization, B is the actual enzyme units that were immobilized on the developed nanosupport.
The kinetic parameters of free and conjugated enzymes were assessed by Lineweaver-Burk plot by monitoring their initial rates at different ONPG concentrations.
Assay of β-galactosidase
Lactase activity assay was performed to hydrolyze β-galactosidase.20 The enzyme activity was determined as the quantity of β-galactosidase that released o-nitrophenol (one micromole per minute) under standard assay conditions.
Lactose hydrolysis in batch process
Free and conjugated enzymes (500 U) were independently added to 100 mM lactose solution (250 ml) under stirring condition in a water bath at 40 and 50 oC for 10 h. Aliquots were collected after a gap of 1 h to monitor the lactose hydrolysis by glucose oxidase-peroxidase assay kit.21
Estimation of protein
Lowry method was used for measuring the protein concentration by using bovine serum albumin as a standard protein.22
Statistical analysis
Sigma software version 9 was used to plot the data. Data was analyzed by one-way ANOVA. All the experiments were performed in triplicates. Probability values <0.05, were considered statistically significant.
Results and Discussion
Importance of the study
β-Galactosidase is extensively used in food and dairy industries for manufacturing lactose-free products and galacto-oligosaccharides, designing of biosensors, bioremediation technology, and for treating biological disorders.23-25 Hence, this was conducted to evaluate lactose conversion in batch reactors by free and conjugated enzymes at 40 and 50 °C.
DLS characterization and steps of immobilization of β-galactosidase
Dynamic light scattering (DLS) confirmed the size of AuNPs as 35 nm (Fig. 1) which agrees with our recently published study.20 The schematic representation of the modification of gold nanoparticles by polyvinyl alcohol followed by the immobilization of enzyme on the modified AuNPs is depicted in Fig 2.
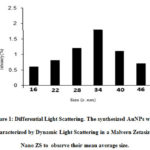 |
Figure 1: Differential Light Scattering. The synthesized AuNPs were characterized by Dynamic Light Scattering in a Malvern Zetasizer Nano ZS to observe their mean average size. |
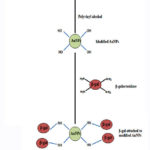 |
Figure 2: Schematic representation of β-galactosidase attached to the modified AnNPs |
Immobilization yield
An enzyme immobilization yield of as high as 88% was obtained as a result of attachment of β-galactosidase on the developed nanomatrix, i.e. PVA-modified AuNPs (Table 1).
Table 1: Immobilization yield. β-galactosidase was mixed with PVA-modified AuNPs as described in the text. Enzyme adsorbed on surface modified AuNPs was washed thrice with assay buffer for calculating the immobilization yield.
|
Loaded enzyme (X Units) |
Enzyme in washes (Y Units) |
Bound enzyme per gram of PVA-modified AuNPs Theoretical (X-Y) = A Actual=B |
Immobilization yield (%) B/A x 100 |
|
1000 |
56 |
944 830 |
88 |
Kinetic parameters measurements
As shown in Table 2, the enzyme immobilization resulted in an increase in Km. However, a decline in Vmax was observed which is attributed to the fact that conjugation resulted in a decline in enzyme affinity for its substrate which ultimately reduced the velocity of enzymatic reaction. The accessibility of substrate was decreased for the active site of the immobilized β-galactosidase which consequently lowered the transport of substrate and products in the developed nanosupport. The obtained results are in compliance with the findings of research conducted on immobilized β-galactosidases from Lactobacillus plantarum HF571129 and Klebsiella oxytoca ZJUH1705 which were exploited for producing galacto-oligosaccharides and lactose hydrolysis.26,27
Table 2: Measurement of kinetic parameters. Km and Vmax of soluble β-galactosidase and enzyme bound to PVA-modified AuNPs were assessed by monitoring their initial rates at different ONPG concentrations.
|
Enzyme |
Km (mM) |
Vmax (mM/min) |
|
Soluble β-galactosidase |
3.56±2.9 |
2.77±3.5 |
|
Enzyme immobilized on PVA-modified AuNPs
|
3.74±1.8 |
2.07±2.7 |
Lactose hydrolysis in batch process
The batch process conversion of lactose solution showed greater hydrolysis (hydrolysis rate) by soluble β-galactosidases initially in contrast to the immobilized enzyme (Table 3). This phenomenon is explained by enzyme mechanics which suggests the easier accessibility of free enzyme that promotes the conversion of lactose into its basic components initially. However, a sudden decrease in the rate of lactose hydrolysis was noticed after prolonged time intervals as a result of product mediated inhibition of β-galactosidase.28 Panesar and co-workers obtained 39% lactose hydrolysis in 4 h at 50 oC by soluble β-galactosidase, however, the greatest percentage of lactose hydrolysis that was accomplished by this system was 71% after 9 h. It was also demonstrated that 83 % of whey lactose could be hydrolyzed by the developed immobilized enzyme after 8 h and under similar experimental conditions. The results obtained above are explained by the fact that the conversion of whey lactose is subjected to the enzyme activity which is based on optimum conditions of β-galactosidase like temperature and pH, and also on the processing time and enzyme concentration.21,29
The present study demonstrated the excellent application of β-galactosidase conjugated to PVA-AuNPs in lactose conversion at 50 oC, owing to its profound thermal stability at this temperature. The fact that β-galactosidases from plant and fungal sources possess acidic pH-optima was discussed in detail.24 Hence, they are suitably exploited for manufacturing acid whey and whey permeates. Contrastingly, β-galactosidases from bacteria and yeasts displayed neutral pH-optima which make them excellent candidates for milk processing. Based upon these findings, researchers obtained 96% lactose from β-galactosidase from Kluveromyces fragilis in 48 h at 35 °C.30 Similarly, Zhou and Chen reported 70% lactose hydrolysis in 3 h at 37 oC from Kluveromyces lactis β-galactosidase conjugated to graphite surface by glutaraldehyde as crosslinking reagent; however, only 50% of lactose was converted after 3 h as the temperature was raised to 50 °C.31 Lactose conversion by the fungal source of β-galactosidase, Aspergillus oryzae, was 64% after 1 h when conjugated to concanavalin A-cellulose.32
Table 3: Lactose hydrolysis. Lactose solution was hydrolyzed by soluble and immobilized [1] β-galactosidase in batch process as discussed in the text. Aliquots were collected after a gap of 1 h to monitor hydrolysis by glucose oxidase-peroxidase assay kit.
|
Time (h) |
Lactose hydrolysis (%) |
|||
|
40 oC |
50 oC |
|||
|
Soluble β- galactosidase |
Immobilized β- galactosidase |
Soluble β- galactosidase |
Immobilized β- galactosidase |
|
|
Run |
0 |
0 |
0 |
0 |
|
1 |
40±1.7 |
47±2.3 |
46±1.9 |
55±2.9 |
|
2 |
58±2.3 |
60±3.7 |
52±2.7 |
60±3.1 |
|
3 |
63±3.4 |
65±2.4 |
55±1.8 |
64±2.2 |
|
4 |
66±3.1 |
71±1.1 |
60±3.5 |
68±3.8 |
|
5 |
67±2.9 |
77±2.6 |
64±1.7 |
75±2.4 |
|
6 |
69±1.5 |
81±1.4 |
64±3.1 |
86±3.2 |
|
7 |
71±2.2 |
81±3.5 |
66±2.2 |
90±1.9 |
|
8 |
71±1.6 |
83±3.8 |
66±3.4 |
94±2.6 |
|
9 |
74±3.3 |
87±2.4 |
70±1.7 |
94±2.8 |
|
10 |
74±2.3 |
87±1.8 |
70±3.4 |
94±2.1 |
Conclusion
The current work understates the predisposition of exploiting PVA-modified AuNPs in various biomedical and biotechnological fields. Another major advantage is that the developed modified nanosupport leads to the reuse of β-galactosidase in an efficient and controlled manner. Lactose hydrolysis results obtained by batch processes at different temperatures further ensure that the developed immobilized system can be successfully employed in continuous reactors to obtain lactose-free dairy items.
Acknowledgment
The critical suggestions of Dr. Rukhsana Satar (Assistant Professor in Biochemistry at Ibn Sina Medical College) are gratefully acknowledged by the authors during the preparation of this manuscript.
Conflict of Interest
The authors declare no conflict of interest.
References
- Saqib, S.; Akram, A.; Halim, S.A.; Tassaduq, R.; 3 Biotech 2017, 7(2), 79-89.
CrossRef - Zibrat, N.; Skrt, M.; Jamnik, P.; Acta. Agric. Slov. 2017, 110(1), 258-269.
CrossRef - Ansari, S.A.; Husain, Q.; J. Mol. Cat. B Enz. 2011, 70(4), 119-126.
CrossRef - Bhattacharjee, S.; Sarker, D.; Int. J. Chem. Eng. Res. 2017, 9(1), 223228.
- Ansari, S.A.; Ahmad, S.I.; Jafri, M.A.; Naseer, M.I.; Satar, R.; Quim. Nov. 2018, 41(2), 429-433.
- Pan, C.; Hu, B.; Li, W.; Sun, Y.; J. Mol. Catal. B Enzym. 2009, 61(3), 208-215.
CrossRef - Ansari, S.A.; Satar, R.; Zaidi, S.K.; Naseer, M.I.; Karim, S.; Alqahtani, M.H.; Rasool, M. Food Bioprod. Proc. 2015, 95(4), 298-303.
CrossRef - Garcia, G.C.; Berenguer, M.A.; Lafuente, R.F.; Rodrigues, R.C.; Adv. Synth. Catal. 2011, 353(4), 2885-2904.
CrossRef - Cipolatti, E.P.; Manoel, E.A.; Lafuente, R.F.; Freire, D.M.G.; Biotechnol. Res. Innov. 2017, 1(2), 26-34.
CrossRef - Cipolatti, E.P.; Silva, M.J.; Klein, M.; Feddern, V.; Feltes, M.M.C.; Oliveira, J.V.; Ninow, J.L.; de Oliveira, D.; J. Mol. Catal. B Enz. 2014, 99(2), 56-67.
CrossRef - Cipolatti, E.P.; Valerio, A.; Henriques, R.; Moritz, D.E.; Ninow, J.K.; Freire, D.M.G.; Manoel, E.A.; Lafuente, R.F.; de Oliveira, D.; RSC Adv. 2016, 106(1), 104675-104692.
CrossRef - Mokhtar, N.M.; Rahman, R.N.Z.R.; Noor, N.D.M.; Shariff, F.M.; Ali, M.S.M.; Catalysts 2020, 10(4), 744-761.
CrossRef - Zdarta, J.; Meyer, A.S.; Jesionowski, T.; Pinelo, M.A.; Catalysts 2018, 8(3), 92-100.
CrossRef - An, J.; Li, G.; Zhang, Y.; Zhang, T.; Liu, X.; Gao, F.; Peng, M.; He, Y.; Fan, H.; Catalysts 2020, 10(2), 338-353.
CrossRef - Ealias, A.M.; Saravanakumar, M.P.; IOP Conf. Ser. Mater. Sci. Eng. 2017, 263(3): 1-15.
- Kockmann, A.; Porsiel, J.C.; Saadat, R.; Garnweitner, G.; RSC Adv. 2018, 8(4), 11109-11118.
CrossRef - Studart, A.R.; Amstad, E.; Gauckler, L.J.; Langmuir 2007, 23(4), 1081-1090.
CrossRef - Cabuzu, D.; Cirja, A.; Puiu, R.; Grumezescu, A.M.; Curr. Top. Med. Chem. 2015, 15(1), 1605-1613.
CrossRef - Chen, Y.; Xianyu, Y.; Jiang, X.; Acc. Chem. Res. 2017, 50(3), 310-319.
CrossRef - Alshanberi, A.M.; Satar, R.; Ansari, S.A.; Molecules 2021, 26(2), 1226-1236.
CrossRef - Ansari, S.A.; Al‐Shaeri, M.; Braz. J. Chem. Eng. 2019, 36(4), 109-115.
CrossRef - Lowry, O.H.; Rosbrough, N.; Farr, A.L.; J. Biol. Chem. 1951, 193(3), 265-275.
CrossRef - Ansari, S.A.; Husain, Q.; Biotechnol. Adv. 2011, 30(2), 512-523.
CrossRef - Husain, Q.; Crit. Rev. Biotechnol. 2010, 30(1), 41-62.
CrossRef - Grosova, Z.; Rosenberg, M.; Rebros, M.; Czech J. Food Sc. 2008, 26(4), 1-14.
CrossRef - Selvarajan, E.; Mohanasrinivasan, V.; J. Food Sci. Technol. 2015, 52(3), 6206-6217.
CrossRef - Huang, J.; Zhu, S.; Zhao, L.; Chen, L.; Du, M.; Zhang, C.; Yang, S.T.; App. Microb. Biotechnol. 2020, 104(2), 6161-6172.
CrossRef - Ansari, S.A.; Husain, Q.; J. Mol. Cat. B Enz. 2011, 70(1), 119-126.
CrossRef - Panesar, R.; Panesar, P.S.; Singh, R.S.; Kennedy, J.F.; Bera, M.B.; Food Chem. 2007, 101(1), 786-790.
CrossRef - Szczodrak, J.A.; J. Mol. Catal. B Enz. 2000, 10(2), 631-637.
CrossRef - Zhou, Q.Z.K.; Chen, X.D.; Biochem. Eng. J. 2001, 9(3), 33-40.
- Ansari, S.A.; Husain, Q.; J. Mol. Catal. B Enz. 2010, 63(4), 68-74.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.










