Novel N2O2 Schiff Base Derived from 1,2-Hydrazinedicarboximidamide and its Complexes with Cu(II), Co(II), Ni(II) , Mn(II) and Cr(III): Synthesis and Characterization
Sara K. Yassin, Jasim M. S. Alshawi* and Zainab A. Mohammedsalih
Basrah – Iraq University of Basrah College of Education for Pure Sciences Chemistry Department.
Corresponding Author E-mail: jasim.salih@uobasrah.edu.iq
DOI : http://dx.doi.org/10.13005/ojc/360520
Article Received on : 04-09-2020
Article Accepted on : 20-10-2020
The synthesis of a (1,2-Hydrazinedicarboximidamide) was identified in this paper and condensing it with 2-4dihydroxybenzaldehyde to form tetradentate ligand (L). This ligand used to prepare five metal complexes as chloride salts [Cu2(L)Cl2](1), [Co2(L)Cl2] (2), [Ni2(L)Cl2](3), [Mn2(L)Cl2](4) and [Cr2(L)Cl2](5) in an ethanolic medium. Dimethyl formamide (DMF) prepared complexes solutions to applied it as electrolytes. The Jasim were confirmed by several spectroscopic and analytical techniques indicating that metal complexes are more likely to have tetrahedral-coordinated geometry. Thus, these structure indicated the ligand show similar actions as tetradentate linked to metal ion by nitrogen (azomethine) and the negative charge of oxygen atoms from hydroxyl in 2-4dihydroxybenzaldehyde.
KEYWORDS:1-2-Hydrazinedicarboximidamide; Complexes; Schiff Base; Tetradentate
Download this article as:| Copy the following to cite this article: Yassin S. K, Alshawi J. M. S, Mohammedsalih Z. A. Novel N2O2 Schiff Base Derived from 1,2-Hydrazinedicarboximidamide and its Complexes with Cu(II), Co(II), Ni(II) , Mn(II) and Cr(III): Synthesis and Characterization. Orient J Chem 2020;36(5). |
| Copy the following to cite this URL: Yassin S. K, Alshawi J. M. S, Mohammedsalih Z. A. Novel N2O2 Schiff Base Derived from 1,2-Hydrazinedicarboximidamide and its Complexes with Cu(II), Co(II), Ni(II) , Mn(II) and Cr(III): Synthesis and Characterization. Orient J Chem 2020;36(5). Available from: https://bit.ly/37OMIRk |
Introduction
Metal complexes of azomethine ligands have acquired more attention due to their easy synthesis, their stereo-electronic structures, and outstanding biological activities (1), catalytic activities (2,3), Synthesis and design of transition metal ion complexes get together. Continued interest of Schiff base ligands consideration of the possibility of earning multiple structures scientific research involving catalysts (4), material science (5), optic applications(6), medical chemistry(7). Synthetic ingenuity leading to variety in structural arrangement in space considered as the gateway factors, which attending wide deal of benefit to azomethane ligands in coordination chemistry. Furthermore, their individual self-assembling behavior, result to the modeling of different supramolecular structural design, have assisted them to have the status of special ligand configuration (8). Schiff bases are believed to be “privileged ligands” due to their easily synthesis by the reaction between aldehyde and primary amine. The Ligands of Schiff base are eligible to linked with several metal ions, and to confirm them in many oxidation states(9). Tetra-dentate Schiff bases with a (N2O2) donation atoms to coordinate with several metal ions, and this has confirmed by many papers(10).
Rather significant advantage was the azomethine group (C = N) in the configuration of the Schiff bases for the lone pair of electrons on a nitrogen atom sp2-hybridized orbital(11). They show premium benefits for several fields, such as speeding up the organic synthesis reaction as a catalyst, chemistry of transition metals, industrial, biological, pharmacological and optical sensor properties (12).
Materials and Methods
Synthesis of (1,2-Hydrazinedicarboximidamide)(A)
It was prepared by reaction of thiourea (20 gm, 0.3228 mol) and hydrazine hydrate (8 gm, 0.16 mol) in ethanol (100 cm3) by refluxing for 3 hours with stirring.The product was cooled at room temperature and filter it to get hydrazide(13). The recommended structure for the synthesis ligand (A) was given in Figure (1) and some physical properties were showed in Tables (1).
2.3. Synthesis of the Schiff Base Ligand (L)
L was synthesis by dissolving (0.039 mol, 5.52g) of 2-4dihydroxybenzaldehyde in (25 ml) of ethanol and adding about (3-2) drops of glacial acetic acid and stirred for 10 minutes to ensure the formation of the carbonium ion, then (0.0096 mol, 1.12g) of compound A dissolved in (15ml) of ethanol was slowly and step by step added while refluxing. After completing the addition process, the mixture left for (5 hr) in an inert atmosphere of Akron gas. The reaction was followed up using layer chromatography(14). The suggested structure for the prepared ligand(L) was showed in Figure (2) and some physical properties were listed in Tables (1).
Synthesis of the Schiff Base Complexes
Complexes of the ions Mn (II), Co (II), Ni (II), Cu (II), Cr (III) were prepared with the Schiff bases of (L) by dissolving (0.149g, 0.001mol) of ligand (Dissolved L in (2: 3: 3 ml) of solvents (DMF: EtOH: MeOH)) respectively with (0.0989, 0.1189, 0.0852, 0.1188, 0.1332g, 0.002 (mol of salts), CuCl2.2H2O CoCl2.6H2O, MnCl2 .4H2O (CrCl3.6H2O, NiCl2.6H2O, respectively, and the mixture still reflux for about (3hr) .With the presence of supplying of Argon gas to the reaction, a colored precipitate was observed after which the solution was filtered and the crystals were washed with diethyl ether and then the product was allowed to dry at room temperature(15). The prepared Complexes structure was suggested in Figure (3) and some physical feature were listed in Tables (1).
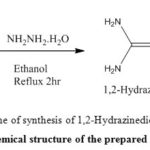 |
Figure 1: Chemical structure of the prepared compound (A) |
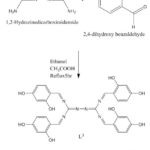 |
Figure 2: Chemical structure of the prepared Schiff base ligand |
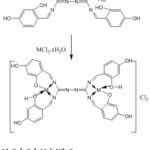 |
Figure 3: Chemical structure of the prepared Schiff Base complexes |
Table 1: Some physical properties of prepared Schiff base and complexes
|
symbol |
Molecular formula |
Molecular weight |
Physical state and colour |
Time of reaction (hour) |
Melting point |
Values Rf |
Yield % |
|
|
A |
C2H8N6 |
116.13 |
White powder |
3 |
139-140 |
0.25 |
58 |
|
|
L3 |
C30H24N6O8 |
596.56 |
Orange powder |
5 |
278-280 |
0.36 |
54 |
|
|
L3-Cu |
[Cu2(C30H24N6O8)]Cl2 |
792.5 |
Brown red crystale |
3 |
˃300 |
—– |
84 |
|
|
L3-Co |
[Co2(C30H24N6O8)]Cl2 |
783.3 |
Olive powder |
3 |
˃300 |
—– |
61 |
|
|
L3-Mn |
[Mn2(C30H24N6O8)]Cl2 |
775.3 |
Yellowish orande powder |
3 |
˃300 |
—– |
49 |
|
|
L3-Ni |
[Ni2(C30H24N6O8)]Cl2 |
782.8 |
Yellowish red powder |
3 |
˃300 |
—- |
63 |
|
|
L3-Cr |
[Cr2(C30H24N6O8)]Cl2 |
769.4 |
Brown crystale |
3 |
˃300 |
—- |
54 |
|
Results
Identification and characterization of synthetic Schiff base and it’s complexes were confirmed by many techniques like by the FTIR,UV-vis, 1H-NMR,13C-NMR,ESI-Mass,molar conductivity and magnetic properties to proven the chemical structure of the prepared Schiff base and it’s complexes.
IR spectra of the ligands and complexes
TheIR bands of the ligands and it’s complexes are given in table (2). The most notation divergence in the IR figure of the ligand (Land complexes (L3-Cu, L3-Co,L3-Mn,L3-Ni,L3-Cr) was the moving of C=N stretching frequencies to smaller frequencies cause to metal– ligand coordination(16). The ligand (L) show vibration of azomethine at 1633 cm-1 was shifted to a greater frequencies in the complex (L-Cu) due to back donation and decreasing the planar specialty after complexation(17).and the azomethine vibration was shifted for a less frequencies in the complexes(L-Co, L-Mn,L-Ni,L-Cr ) due to decreasing the double bonding property after complex (18). The absorption bands in the spectra of complexes(L-Cu) (L-Co), (L-Mn),(L-Ni), and (L-Cr ) at 3363cm-1, 3344cm-1 , (3348-3336cm-1), 3367cm-1 and 3385 cm-1 respectively were attributed to OH.
Table 2: IR bonds (cm-1) of the ligands and the complexes
|
symbol |
C-H (Ar.)
|
C=N
|
C=C (Ar.)
|
Other
|
N-N
|
C-O |
C-N |
M-N |
M-NH |
M-O |
|
A |
—– |
1643-1612 |
—— |
N-H (3377-3174) |
1083 |
—— |
1473-1415 |
—– |
—– |
—- |
|
L |
3039 |
1633 |
1581-1444 |
O-H 3130 |
1166 |
1230 |
1396 |
—— |
—– |
—— |
|
Cu L)) |
3120 |
1643 |
1537-1487 |
O-H 3363 |
1122 |
1228 |
1396 |
412 |
— |
497 |
|
L) Co) |
3205 |
1616 |
1541 |
O-H 3344 |
1110 |
1222 |
1435 |
462 |
—- |
516 |
|
(Mn L) |
3215-3203 |
1618 |
1498 |
O-H 3348-3336 |
1162 |
1230 |
1325 |
499 |
—– |
634 |
|
(Ni L) |
3184 |
1631-1618 |
1585-1498 |
O-H 3367 |
1130 |
1230 |
1395 |
511 |
—– |
634 |
|
Cr L)) |
3132-3066 |
1618 |
1583-1539 |
O-H 3385 |
1133 |
1234 |
1429 |
480 |
—– |
426 |
UV– Visible spectra of the ligands and complexes
The electronic spectra (UV/vis) data of the ligands and its complexes are shown in table (3) and The π band of the ligands Lat 282 nm is assigned to phenyl of 2,4-dihydroxybenzaldehyde π→π* transition(19). The azomethine group in these ligands was appeared two bands is because of the π→π* and n→π*(19). These two bands were moved to higher wavelengths in the complexes. The spectra of complexes (L-Cu, L-Co, L-Mn, L-Ni, L-Cr) show new two types of bands in higher wavelengths (visible region) are indicated to d–d transitions of the metal ion (22) and ligand – metal charge transfer (LMCT) bands (20).
Table 3: Electronic spectral data for the ligand and their complexes
|
Symbol compound |
Wavelength and molarabsorption ( ) |
Wavelength (n ) |
Wavelength and molar absorption (L-M) |
Wavelength and molar absorption (d-d) nm |
Transfer type (d-d) |
structure |
|
L |
282,318,16000,11000 |
356,1630 |
—— |
—— |
—— |
tetrahedral |
|
Cu L)) |
286 1620 |
358 504 |
440 78 |
518 37 |
3T2→3E |
tetrahedral |
|
L) Co) |
284,320 1650,1090 |
—– |
376 80 |
520,725,50,40 |
4FA2→4PT1 4FA2→4T2 |
tetrahedral |
|
(Mn L) |
259,279,324, 1647,2060, 1550 |
—— |
—— |
481,730,42,15 |
Tetrahedral |
|
|
(Ni L) |
282 1380 |
321 890 |
380 112 |
490 3.8 |
3FT1→3PT1 3FT1→3A2 3FT1→3T2 |
tetrahedral |
|
Cr L)) |
297,322 740,850 |
328 374 |
——- |
460,600, 774,220,23,24 |
4FA2→4PT1 4FA2→4T2 |
tetrahedral |
1H-NMR, 13C – NMR spectra of the ligands
The 1H– NMR spectra of the ligand L was recorded in DMSO. Using 400 MHZ, and it appeared a many of characteristic signals of the ligand as shown in table (4). The signals due to the aromatic protons were observed in the range 𝛿 = 7.53 – 6.32 ppm. The signal observed in the 𝛿 = 9.90 ppm was attributed to the azomethine protons in the molecule(21). 13C – NMR spectra of the ligand was recorded in DMSO. Using 100MHZ ,and it showed a number of identification signals of ligand as shown in table (5)(17), the signals assigned the aromatic carbon were shown in the range .the signal observed in the 𝜹165.16ppm was due to azomethine carbon in the molecule and the signals observed in 𝜹163.25 ppm was attributed to carbon guanidine group (22).
Table 4: Chemical shift of 1H-NMR for the ligand(L3)
|
Symbol compound |
protons |
signal |
(chemical shift ppm) |
|
L |
1H,HC=N |
singlet |
9.90 |
|
3H,Ar-H |
multiple |
7.53-6.32 |
Table 5: Chemical shift of 13C-NMR for the ligand(L3)
|
Carbon atoms |
(chemical shift ppm) |
|
L3 |
|
|
C1,(-C=N-N) |
163.25 |
|
C2,(HC=N) |
165.16 |
|
Ar-c |
102.19-132.81 |
ESI-Mass Spectra
ESI-Mass spectra of the synthesised compounds were carried out by using Agilent Technologies–Tehran-Iran-Trabiat Modares University. ESI-Mass spectra of the prepared ligand and complexes showed molecular ion peak (M+) at 116,which corresponding to the molecular formula of (A), at 733 which corresponds to the molecular formula of (L).And at (723),(713),(704),(714),(701) which corresponds to molecular formula of complexes (L-Cu), (L-Co), (L-Mn), (L-Ni), (L-Cr)(23). ESI-Mass spectra of the synthesised compounds are shown in table (6)and Figs (18 – 24)
Table 6: ESI-Mass spectra of prepared ligands and complexes
|
Relative abundance |
M+ |
Molecular weight |
Molecular formula |
compound |
|
83 |
M=116 |
116.13 |
C2H8N6 |
A |
|
157 |
M+2H=599 |
596.56 |
C30H24N6O8 |
L |
|
341 |
M+H=723 |
721.62 |
C30H24N6O8)Cl2)Cu2 |
Cu L)) |
|
203 |
M+H=713 |
712.39 |
C30H24N6O8)Cl2)Co2 |
L) Co) |
|
153 |
M=704 |
704.4 |
C30H24N6O8)Cl2)Mn2 |
(Mn L) |
|
143 |
M+2H=714 |
711.9 |
C30H24N6O8)Cl2)Ni2 |
(Ni L) |
|
203 |
M+2H=701 |
698.52 |
C30H24N6O8)Cl2)Cr2 |
Cr L)) |
Molar Conductivity
The measurements of Conductivity were completed at room temperature of 10-3 M in DMF solvent to proven the charge order of metal complexes. The values of conductivity for the prepared metal complexes Co (II), Cu (II) , Mn(II), Ni(II) and Cr(III) were found to be 82.1, 80.2, 89.5, 79.7and 78.2 Ohm-1 cm2 mol-1 respectively. So that, the suggestion for the complexes were electrolytes (24) due to the Chlorate ions outside the coordination sphere in solution and they were separate gradually in the DMF solvent.
Table 7: Conductivity of complexes
|
Molar conductivity Ohm-1.cm2.mol-1 |
Electrical conductivity Ohm-1 |
Cell constant A(cm-1) |
Molecular formula |
Symbol compound |
|
82.1 |
82.1˟10-6 |
1.1 |
[Cu2(C30H22N6O8)]Cl2 |
Cu L)) |
|
80.2 |
80.2˟10-6 |
1.1 |
[Co2(C30H22N6O8)]Cl2 |
L) Co) |
|
89.5 |
89.5˟10-6 |
1.1 |
[Mn2(C30H22N6O8)]Cl2 |
(Mn L) |
|
79.7 |
79.7˟10-6 |
1.1 |
[Ni2(C30H22N6O8)]Cl2 |
(Ni L) |
|
78.2 |
78.2˟10-6 |
1.1 |
[Cr2(C30H22N6O8)]Cl2 |
Cr L)) |
Magnetic Susceptibility Measurements
The calculated of magnetic moment to prepared complexes displayed that the d9 Cu(II) complex is 1.843.6BM reported as distorted tetrahedral geometry. The observed of magnetic moment to the d7 Co(II) complex is 2.531BM which is more corresponding with the SP3. The magnetic susceptibility measurement determined for the d5 Mn(II) complex is 5.836BM which suitable with the distorted tetrahedral geometry . The magnetic moment showed for d8 Ni(II) complex is 3.463BM which is more convenient with the SP3 geometry while Cr Complex computed 3.932 BM which is more convenient with the tetrahedral geometry(25)too.
Table 8: Magnetic susceptibility measurements
|
Hybridization |
Effective magnetic moment Μeff(B.M.) |
Atomic Magnetic Susceptibility XA.10-6 |
Correct Factor D.10-6 |
Molar magnetic susceptibility Xm.10-6 |
Mass magnetic susceptibility Xg.10-6 |
Symbol of complex |
|
Tetrahedral |
1.8436 |
1600.36 |
379.3- |
1979.66 |
2.498 |
Cu L)) |
|
Tetrahedral |
2.531 |
3437.7 |
380.1- |
3817.80 |
4.874 |
L) Co) |
|
Tetrahedral |
5.836 |
5040.72 |
382.5- |
5423.22 |
6.995 |
(Mn L) |
|
Tetrahedral |
3.463 |
4228.42 |
371.9- |
4600.32 |
5.873 |
(Ni L) |
|
Tetrahedral |
3.932 |
3386.76 |
385.6- |
3772.36 |
4.903 |
Cr L)) |
Discussion
The L ligand and its metal complexes of Cu(II), Co(II), Mn(II), Ni(II) and Cr(II) have been structurally proven by several techniques . The stoichiometry of metal ligand in all these complexes clarify that analytical data is 2:1. In DMF solvent appeared all these complexes are electrolytes. That ligand showed the spectral data deal as neutral and tetradentate coordinating through nitrogen atom of the azomethine and oxygen atoms of hydroxyl group of the 2-4dihydroxybenzaldehyde .Depending on all information that gotten from technique like molar conductance, magnetic and spectral data, lead to considerable opinion that complexes wer assigned to be in tetrahedral geometry (SP3).
Acknowledgement
The authors are grateful to all lab mate for corporation.
Conflict of Interest
The authors declared that there is no conflict of interest regarding the publication of this paper.
References
-
Hajrezaie M., Paydar M., Looi C. Y., Moghadamtousi S. Z., Hassandarvish p., Salga M. S., Karimian H., Shams K., Zahedifard M., Majid N. A., Ali H. M., Abdulla M. A., Sci.Rep., 2015, 5, 9097.
CrossRef -
Alzoubi W., Ko Y.G., J.Organomet.Chem., 2016, 822 , 173.
CrossRef -
Gong D., Wang B., Jia X., Zhang X., Dalton Trans., 2014, 43, 4169.
CrossRef -
Kwong H.L., Yeung H.L.,Yeung C.T., Lee W.S.,Lee C.S.,Wong W.L., Coord.Chem.Rev., 2007,251, 2188-2222.
CrossRef -
Vigato P.A., Tamburini S., Coord.Chem.Rev., 2004, 248, 1717-2128.
CrossRef -
Wanga H., Hossain A.M.S.,Zhang Q., Wua J., Tian y.,Inorg. Chim. Acta., 2014, 414, 153-159.
CrossRef -
Bagherzadeh M., Amini M., Ellern A., Woob L.K., Inorg.Chim. Acta., 2012, 383, 46-51.
CrossRef -
Biswal D., Pramanik N.R., Chakrabarti S., Chakraborty N., Acharya K., Mandal S.S., Ghosh S., Drew M.G.B., Mondale T.K., Biswas S., New J.Chem., 2015, 39, 2778-2794.
CrossRef -
Sunatsuki Y., Motoda Y., Matsumoto N., Coord.Chem.Rev., 2002, 226, 199-209.
CrossRef -
Gaeg B.S., Kumar D., Spectrochim. Acta A. Mol. Biomol. Spectrosc., 2003, 59, 229-234.
-
Layek S., Agrahari B., Tarafdar A., Kumari C., Ganguly,D. D., Pathak A.R., J. Mol. Struct., 2017, 428, 1141.
CrossRef -
Sirajuddin M., Uddin N., Ali S., Nawaz T.M., Spectro chim.Acta part A, 2013, 116,111.
CrossRef -
EL-Tabl A.S., Stephanos J. J., Abd-Elwahed M. M., Metwally E. E. D. and El-Gamasy S. M., J.Chem. Bio. Phy. Sci. Sec .A, 2015, 5, 3875-3908.
-
M.Sondhi S., Dinodia M., Jain S. and Kumar A., Indian J.Chem ., 2009, 48B, 1128-1136.
-
Mohammed N.L., AL-Shawi J. M. S. and Kadhim M.J., Int. J. Sci. Eng. Res., 2019, 7, , 31-40.
-
Giesbrecht E., Pure Appl. Chem, 1979, 51, 925-947.
CrossRef -
Gokhale N. H., Padhye S. S. and Padhye S. B., Inorganica Chim. Acta, 2001, 319, 90-94.
CrossRef -
Silversten R. M., Webster F. X., Kiemle D. J., Spectrometric I dentification of Organic Compound,7th Ed, John Wiley & Sons Ltd, United Kingdom, 2005.
-
Kaczmarek M. T., Skrobanska M., Zabiszak M., Watesa-Chorab M., Kubicki M. and Jastrzab R., Royal Society of Chemistry, 2018, 8, 30994.
CrossRef -
Andiappan K., Sanmugam A., Deivanayagam E., Karuppasamy K., kim H. and Vikraman D., Int. J. Biol. Macromolecules, 2019, 124, 403-410.
CrossRef -
Gupta S. D., Revathi B., Mazaira G. I., Galigniana M. D., Subrahmanyam C. V. S., Gowrishankar N. L. and Raghavendra N. M., Bio-organic Chemistry, 59 , 2015, 97-105.
CrossRef -
Jayaprakashr, S. K. Hemalathas S. and Easwaramoorthyd A., Asian J. Pharm. Clin. Res., 2016, 9, 203-208.
-
Gutowska N., Seliger P., Romanski J., Zieba M., Adamus G. and Kowalczuk M., Molecules, 2020, 25(1), 136.
CrossRef -
Geary W.J., Coord. Chem. Rev., 1971, 7, 81-122.
CrossRef -
Cotton F.A. and Wilkinson G., “Advanced Inorganic Chemistry. A Comprehensive Text, 4th ed.; John Wiley: New York, 1980.

This work is licensed under a Creative Commons Attribution 4.0 International License.









