Leucine Aminopeptidase from Arachis hypogaea L. Seeds Partial Purification and Characterization
Taghreed U. Mohammd1 Layla O. Farhan2 Ashgan S. Dawood2 and Bushra F. Hasan2
1Department of Chemistry, College of Education for Pure Sciences, Ibn-AL-Haitham-University of Baghdad, Baghdad,Iraq
2Department of Chemistry, College of Science for Woman, University of Baghdad, Baghdad, Iraq.
DOI : http://dx.doi.org/10.13005/ojc/330567
Leucine aminopeptidases (LAP; EC 3.4.11.1) constitute a diverse set of exopeptidases that catalyze the hydrolysis of leucine residues from the amino-terminal of protein or peptide substrates , (LAP) are present in animals, plants, and microbes . In this study, leucine aminopeptidase was purified Partial from Arachis hypogaea seeds by using gel filtration chromatography Sephadex G-100. The enzyme was purified 3.965 fold with a recovery of 29.4%. Its pH and temperature optimum were(8.7) and (37˚C), respectively. The results show novel properties of LAP from Arachis hypogaeaL. or peanut. The Km value for LAP (77 mM) , with Vmax (1538 mmole min -1). We recommend a separate isoenzyme of the enzyme (LAP)from Arachishypogaeaon L. peanut seeds and study the kinetic qualities of each of them .
KEYWORDS:Leucine amionpeptidase (LAP); Arachis hypoaea L. seeds; Purification
Download this article as:| Copy the following to cite this article: Mohammd T. U, Farhan L. O, Dawood A. S, Hasan B. F. Leucine Aminopeptidase from Arachis hypogaea L. Seeds Partial Purification and Characterization. Orient J Chem 2017;33(5). |
| Copy the following to cite this URL: Mohammd T. U, Farhan L. O, Dawood A. S, Hasan B. F. Leucine Aminopeptidase from Arachis hypogaea L. Seeds Partial Purification and Characterization. Orient J Chem 2017;33(5). Available from: http://www.orientjchem.org/?p=38101 |
Introduction
Arachis hypogaea L. (Peanut) is one of seeds that has great quantity of oil and protein , therefor, it is considered as very important food legume to human nutrition in the world . We can find Arachis hypogaea L. (Peanut) widely distributed in the tropical and subtropical areas of the world [1]. It is an annual herbaceous plant growing (30-50) cm (1.0-1.6) feet tall. It has opposite leaves , pinnate with four leaflets (two opposite pairs; no terminal leaflet). The flowers of Arachis hypogaea L. are similar to typical pea flowers in shape, its color is yellow with reddish veining [2] .
Biochemical, physiological, chemotaxonomy and gentic variability investigation studies make use of qualitative and quantitative isoenzymatic analysis, these studies benefit of a large number of plant population, cultivars and species [3].
Aminopeptidase (EC 3.4.11) is one of the group of proteases, important enzymes that play major role in many different life processes. The hydrolysis of amino acids can be catalyzed by using Aminopeptidase which found in the N-terminal of peptide and are involved in proteins degradation to free amino acids [4]. The major and greatest studied group of aminopeptidase is Leucin aminopeptdase (LAP, EC 3.4.11.1) , especially in microorganisms and animals, therefor recognize it is x-ray crystal structure and their nucleotide sequences [5].
Generally, two classes of leucyl aminopeptidases (LAPs) have been reported for most plant species [6]. The first group has thermolabile aminopeptidases with molecular weight of approximately 60-90 KDa and a neutral pH optimum. The second group are enzymes like plant LAPs but isolated from animals. With large (250-330 KDa), homohexameric metallopeptidases that contain ethylene diamine tetraacetic acid (EDTA) and be statin. They are heat stable and possess on alkaline pH optimum. LAPs with alkaline pH optimum have been biochemically purified from a number of plants [7].
In the present study, we purified major LAP from Arachis hypogaea L. seeds and characterized its enzymoloical properties (pH and temperature optimum).
Material and Methods
Enzyme Extraction
Arachis hypogaea L. (Imported from Turkey )was obtained from a local super market in Baghdad, Iraq. Homogenizing 1g homogenate at 18000 g for 30 of Arachis hypogaea seeds in 5mL of phosphate buffer, pH 7.8 use a Teflon pestle homogenizer were used for preparing crude extract. The supernatant was separated as crude after getting rid of insoluble debris by centrifuging min at 4°C .
Detrminations of protein concentration
This method is based on the reaction of proteins with the alkaline sulphate followed by Folin-cicalteau reagent which produces a blue color complex due to the reaction of alkaline copper sulphate with the protein, the intensity of the color depends upon the quantity of the protein detected [8]. As shown in figure (1).
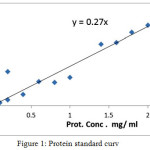 |
Figure 1: Protein standard curve. Click here to View figure |
hydrolysis of the peptide bond of leucinamide is measured according to Mitz and Schlueter method spectrphotometrically at 238 nm .Unit of enzyme activity like one micromole of L- lucinamide hydrolyzed per minute at 25˚C and pH 8.5 [9].
Parital purification of LAP
Gel filtration chromatography using sephadex G-100 column was used for purification of partially purified enzyme. The height of 120 cm in a glass column with an internal diameter of 2.0 cm were used for packing the column ,then equilibrated with 0.1 M Tris-HCl buffer pH 8.5 .The flow rate was at 0.5 mL min-1. Forty fractions had been collected for each 2 mL and both the protein content and the enzyme activity were determined for each separate fraction, as pointed out in the previous section.
Effect of pH on LAP Activity
The effect of pH on LAP activity in the purified extract was firmed in different pH (8.1, 8.3, 8.5, 8.7, 8.9) with Tris-HCl buffer pH 8.5. Then LAP activity was measured
Effect of temperature on LAP Activity
The effect of temperature on LAP activity was firmed in different temperature (20, 24, 37, 40, 45˚C) with Tris-HCl buffer pH 8.5. Then LAP activity was measur
Different Concentration of Substrate
Different concentration have been prepared (45, 85, 125 , 165, 205 ) mM /L of substrate (leucinamide) in buffer [9] Km and Vmax for enzyme to substrate were determined by using the Lineweaver-Burk plot [the relationship between 1/V versus 1/[S].
Result and Discussion
Table 1 summaries the result of partial purification of LAP from Arachis hypogaea. The supernatant with LAP activity of 1486.0 Unit/mL and specific activity of 413.23 Unit/mg was considered as crude enzyme solution.
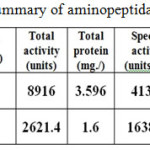 |
Table 1: Purification summary of aminopeptidase Arachis hypogaea. Click here to View table |
This crude enzyme solution fraction was made up to a known volume, partially purified LAP was obtained from this crude enzyme solution fraction and loaded on sephadex G-100 gel filtration column.Gel filtration was purified to the enzyme 3.965 fold with a yield 29.4% and specific activity of 1638.75 Unit/mg. as shown Figure (2).
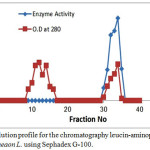 |
Figure 2: Atypical elution profile for the chromatography leucin- aminopeptidase from Arachishypogaeaon L. Click here to View figure |
The exact role of LAPs in plants is not known, while it is enzymes take part in some important processes, such as protein mobilization from cotyledons after germination, and protein turnover required for cell maintenance in vegetative and reproductive organs[10]. LAPs are involved in rapid turnover of protein required in wounding or wounding or pathogen attack [11].
The highest enzyme activity in the range range of pH (8.1 – 8.9), with optimum activity in Tris-HCl buffer at pH 8.7 was shown as in figure (3). This result was agree with the value got from Fasciola gigantic LAPat pH 8.0 [12], from Kiwifruit pH 9.0 [13]. In comparison with , aromatic aminopeptidase which was isolated from grapes gave a result which was pH 7.0 [14], from barley pH7.2 [15] and from wheat pH 7.6 [5]. While Matsui et al 2006 [9] showed that maximum activity was obtained at alkaline pH (8.0-11.0).
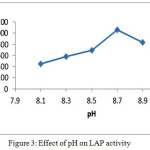 |
Figure 3: Effect of pH on LAP activity. Click here to View figure |
The optimum temperature for the activity of LAP from Arachis hypogaea L. seeds was determined to be 37˚C. as shown in figure (4). This result like the value obtained from pea shoots 37˚C [16,17]. In the other studies kiwifruit AP was most active at 37˚C [13]. Unlike stability of aromatic aminopeptidase, lucine aminopeptdases remained even at temperatures over 60˚C, an optimum temperature of 60-70 ˚C was used for characterizing their activities [9]
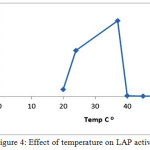 |
Figure 4: Effect of temperature on LAP activity. Click here to View figure |
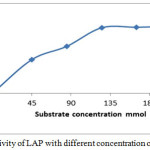 |
Figure 5: Activity of LAP with different concentration of substrate . Click here to View figure |
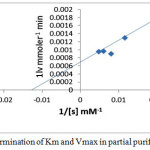 |
Figure 6: Determination of Km and Vmax in partial purified enzyme. Click here to View figure |
A linear relationship was obtained a Vmax [1538 U/mL] and Km value of [77 mM].An enzyme with low Km has a more affinity for its substrate .
Conclusion
In conclusion , in the present study,it was found one peak of the Arachishypogaeaon L. peanut enzyme (LAP) by chromatography(gel filtration ).The Arachishypogaeaon L. peanut enzyme (LAP) reach optimum activity at pH is 8.7 and temperature at 37 o C . The purified enzyme indicated maximum activity(Vmax) of 1538 m mole min -1 with its parallel Km value of 77 Mm .We recommend a separate isoenzyme of the enzyme (LAP)from Arachishypogaeaon L. peanut seeds and study the kinetic qualities of each of them . Note that the enzyme was purified for the first time and have been a successful and appropriate way because we get a good find yield.
References
- Maria, L., and Catalina. R.L. 1994 . Isoenzimatic variability among five peanut cultivars.Bragantia.Campinas.,53(2):135-140.
- Ryals , J.; Ward, E.; Ahl-Goy, P.,and Metraux, J. P. 1992 . Inducible plant proteins: Society for Expermintal Biology Series, ed. Wary, J. L. (Cambridge Univ. Press, New York), 49 :205-229.
CrossRef - Cherry, J.P., and Ory, R.L. 1973. Electrophoretic characterization of six selected enzymes of peanut cultivars. Phytochemistry, Oxford, 12(7):283-289.
CrossRef - Joana, K., and Danuta. G. 2015 . Aminopeptidases isolated from plants of great economic value-role and characteristics. Chemik., 69(8): 463-468.
- Magda, P.; Urszula, S.;Wieslaw, B. and Edyta, Z. 2011. Purification, biochemical characterization, and mass spectrometry analysis of phenylalanine aminopeptidase from the shoots of pea plants. Acta Physiol Plant., 33:609-617.
CrossRef - Zoran, V.; Bilijana, D.; Aleksandra, M. and Natasa, B. 2008 . Purification and Properties of the Major Lucyl Aminopeptidase from Solanum tuberosum Tubers. Fruit, Vegetable and Cereal Science and Biotechnology. Special Issue 1., 125-130.
- Matsui, M.; Fowler, HJ. and Walling, LL. 2006. Lucine aminopeptidase: diversity in structure and function. Biological Chemistry., 387: 1535-1544.
CrossRef - Sadasivam, S. and Manikam, A. 1992 . In: Biochemical Methods for Agricultural Science, Wily Eastern Limited, New Delhi and TNAU, Coimbatore., India, p:122-123.
- Mitz, M. and Schlueter, R.1958. Direct Spectrophotometric Measurement of the Peptide Bond: Application to the Determination of Acylase 1. Biochim. Biophys. Acta, 27:168.
CrossRef - Park, SY. And Walling, LL. 2003 . Isolation and characterization of the neutral leucine aminopeptidase (LapN) of tomato. Plant Physiology., 132(1):243-255.
CrossRef - Gu, YQ. and Walling, LL. 2000. Specificity of the wound- induced leucine aminopeptdase (LAP-A) of tomato activity on dipeptide and tripeptide substrates. European Journal of Biochemistry,207(4):1178-1187.
CrossRef - Daniel, C.; Fernando G. and Carlos, C. 1998. Characterizayion and Partial Purification of a Leucine aminopeptidase From Fasciola Hepatica. J. Parasitol., 84(1):1-7.
CrossRef - Permarathne, A.A. and David, W.M. 2010. Characterization of Activity of a Potential Food-Grade Lucine Aminopeptidase from Kiwifruit. Enzyme Research, Articale ID 517283:1-5.
- Mahagamasekera, M.G.P. and Leung, W.M. 2001. Development of leucine aminopeptidase activity during dalily flower growth and senescence. Acta Physiologiae Plantarum, 23(2): 181-186.
CrossRef - Kang, H. C.; Hahn,T.-R.; Chung,I-S. and Park, J-C. 1999. Characterization of aminopeptidase from grapes. Int J Plant Sci., 160:299-306.
CrossRef - Oszywa,B.; Makowski, M. and Pawelczak, M. 2013. Purification and partial characterization of aminopeptidase from barley(Hordeum vulgare L.)seeds. Plant Physiol. Bioch., 65:75-80.
CrossRef - Ogiwara, N.; Amano, T.; Satoh, M. and Shioi, Y. 2005. Lucine aminopeptidase from etiolated barley seedling: characterization and partial purification of isoforms. Plant Sci., 168:575-581.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









