Isolation of Lupeol, Design and Synthesis of Lupeol Derivatives and their Biological Activity
S. Devendra Rao1, B. Nageswara Rao1, P. Uma Devi2 and A. Karteek Rao2
1Divi’s laboratories limited, Unit-II, Chippada, Visakhapatnam, Andhra Pradesh, India-531162.
2Department of Chemistry, GIS, GITAM University, Rushikonda, Visakhapatnam, Andhra Pradesh, India-530 045.
DOI : http://dx.doi.org/10.13005/ojc/330119
Article Received on :
Article Accepted on :
Article Published : 15 Feb 2017
The triterpenoid, lupeol (1) has been isolated from the leaves extract of Walsura trifoliate. Few novel derivatives (4a–j) were synthesized from the naturally occurring lupeol (1) and confirmed by spectroscopic methods, and tested for antimicrobial and anti-proliferative activity against MDAMB-231, IMR32and A549 cell lines. This all compound showed moderate activities
KEYWORDS:Walsura trifoliata; Lupeol; oxime ester derivatives
Download this article as:| Copy the following to cite this article: Rao S. D, Rao B. N, Devi P. U, Rao A. K. Isolation of Lupeol, Design and Synthesis of Lupeol derivatives and their Biological activity. Orient J Chem 2017;33(1). |
| Copy the following to cite this URL: Rao S. D, Rao B. N, Devi P. U, Rao A. K. Isolation of Lupeol, Design and Synthesis of Lupeol derivatives and their Biological activity. Orient J Chem 2017;33(1). Orient J Chem 2017;33(1). Available from: http://www.orientjchem.org/?p=30199 |
Introduction
Triterpenoids are secondary metabolites derived from plants, fungi etc. triterpenoids contain about thirty carbon atoms, and their structures are considered to be derived from the acyclic precursor squalene 1. Triterpenes are widely distributed in nature; more than 20,000 triterpenoids has been isolated from Nature, which belong to different groups such as squalene, lanostane, dammarane, lupane, oleanane, ursane, hopane, etc.2. Among these triterpenoids, lupane has attracted much attention due to their broad spectrum of biological activities.
The genus Walsura Roxb. (Meliaceae) is a rich source of triterpenoids3 with promising biological activities. Walsura trifoliata (synonym: Walsura piscidia Roxb.) is distributed widely in the tropical areas of China, India, Malaysia, and Indonesia. These are well known for their medicinal properties like treatment of skin allergies, astringency antimicrobial activity and diarrhea 4. Walsura trifoliate is a new source of Lupeol and we first time isolated from leaves through chloroform extract.
Lupeol (1) is a lupane type pentacyclic triterpene is principally found in common fruit plants such as olive, mango, plant, fungi etc.5. Lupeol exhibit an array of pharmacological activities against various diseases include anti-inflammation 6, anti-arthritis 7 ,anti-oxidant8,9, anti-tuberculosis10, diabetes, cardiovascular ailments, anti HIV11, hepatic toxicity, microbial infections and cancer12. The literature also suggests that lupeol significantly reduced the growth of human PaC tumors and regulates over expression of cellular FLICE-like inhibitory protein (cFLIP)13, modulates NF-jB and PI3K/Akt pathways and inhibits skin cancer14. In recent years there has been rapid progress in the field of derivitization of natural products with the increase in biological activity than the parent molecule. Thus the synthesis of lupeol and its ester on cyclophosphamide-induced hyperlipidaemic cardiomyopathy protection to the cardiac tissue , could preserve membrane permeability15 could preserve lysosomal integrity, improve thiol levels16. Lupeol and their ester derivatives show anti-diabetic17, anticancer, anti-inflammatory18, hypercholesterolemia19 etc. Very few oxime derivatives have been reported. This motivated us to study the biological properties of oxime ester derivatives of lupeol.
By analyzing Lupeol, the OH group at C-3 position can be modified for the synthesis of oxime derivatives. In continuation of our interest on synthesis of derivatives of biologically active natural products, we report synthesis of series of oxime ester derivatives (4a to j) from natural product Lupeol.
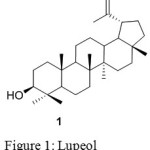 |
Figure 1: Lupeol Click here to View figure |
Materials and Methods
Experimental
All the solvents were dried by using drying agents and distilled prior to use. The reagents were purchased from Aldrich and Across and were used without purification unless otherwise stated. All moisture-sensitive reactions were carried out under nitrogen condition. Silica gel (Acme 60–120 mesh) was used for Column chromatographic separations. 1H NMR (300 MHz and 500 MHz) and 13C NMR (75 MHz and 125 MHz) spectra were measured with Varian 500 MHz and Bruker Avance 300 MHz with tetramethylsilane as internal standard for solutions in deuteriochloroform. J values are given in Hertz. Mass spectra were recorded on Agilent Technologies 1100 Series (Agilent Chemstations Software).
Isolation of lupeol from Walsura trifoliate
Lupeol was isolated for the first time form Walsura trifoliata. The fresh leaves of Walsura trifoliate were collected and shade dried for 48 hrs and coarsely powdered. About 1 kg powdered leaves was taken into RBF and extracted with chloroform using soxlet apparatus for 12 hrs. The resultant extract was dried and purified using column chromatography.
Synthesis of oxime ester derivatives of Lupeol (4a-j)
Step 1
Synthesis of lupeol ketone intermediate (2)
Exactly weighed 1 gm of Pyridinium chlorochromate (PCC) (2.34 mmol) was added to solution containing 75 mg of lupeol (1) (3.52 mmol) dissolved in 5 mL DCM. Stirr the reaction mixture at room temperature for 3 h and 10 mL of isopropanol was added slowly. The reaction mixture is kept again for stirring continuously at room temperature until the reaction is completed (TLC). After completion of reaction the excess of solvent was removed under reduced pressure. In order to separate the analogues from organic compounds, the residue was triturated with ether and the organic layer was separated. Then the separated organic layer was washed with 1M HCl followed by 10 ml of brine solution. The resulting mixture was concentrated and the crude obtained was purified by column chromatography using EtOAC/Hexane (1:9) to give pure compound (2) (yield: 842 mg, 85%).
Step: 2
Synthesis of lupeol oxime intermediate (3)
Exactly weighed 0.54 gms of above synthesised compound (2) and 0.13 gms of hydroxylamine hydrochloride were taken in 5 ml of dry pyridine and refluxed for 1 hr. the reaction was monitored using TLC. After cooling, the reaction mixture was poured onto crushed ice and extracted using ethyl acetate. The resulting organic layer was concentrated and the crude product was purified by using column chromatography with EtOAC/Hexane (2:8) to give pure compound (3) (Yield: 684 mg, 80%).
Step-3
Synthesis of lupeol oxime ester derivatives
Exactly weighed 20 mg of compound (3), 17 mg of substituted aromatic acids are mixed in 2 mL of dry THF to this mixture 33.56 µml of Et3N was added and stirred for 0.5 h at room temperature. To this solution 17.74 µml of 2, 4, 6-trichlorobenzoyl chloride in 2 mL dry THF was added and stirring was continued for 5 h at room temperature the reaction was monitored by TLC. The solvent was evaporated and the remaining residue was diluted with 2mL of toluene followed by catalytic amount of DMAP and finally with oxime alcohol 3 (660 mg, 1.50 mmol) and stirred for 14 h at room temperature. Toluene was evaporated and crude residue purified by column chromatography EtOAC/Hexane (1:9) to give pure compound KSB-1 (53 mg, 82%).
(1R, 3aR, 5aR, 5bR, 11aR, E)-3a, 5a, 5b, 8, 8, 11a-hexamethyl-1-(prop-1-en-2-yl) octadecahydro-1H-cyclopenta [a] chrysen-9 (5bH)-one O-4-ethylbenzoyl oxime (4a)
Colorless amorphous powder, IR (KBr) cm-1 1633 (C=N), 1556 (C=C), 1H NMR (300 MHz, CDCl3): δ = 7.97 (2H, d, J =8.2 Hz), 7.28(2H, t, J =8.2 Hz), 4.69 (1H, s), 4.57 (1H, s), 2.96-2.90 (1H, m), 2.69 (1H, q, J = 8.2, 7.6, 15.2 Hz ), 2.51 (1H, m), 1.67 (3H, s), 1.40 (3H, s), 1.37 (3H, s), 1.25(3H, s), 1.15 (3H, s), 0.93 (3H, s), 0.79 (3H, s) 2.17(1H, m),1.45 (4H, m), 1.32 (4H, m), 1.29 (1H, m), 1.66 (1H, m), 1.45 (1H, m), 1.38 (1H, m), 1.46 (4H, m), 1.18 (4H, m), 1.54 (2H, m). 13C NMR (75 MHz, CDCl3): δ = 176.3, 164.3, 150.9, 149.7, 129.6, 127.9, 127.1, 109.3, 55.3, 49.9, 47.9, 42.9, 42.8, 41.5, 40.8, 39.9, 39.0, 37.1, 35.4, 33.7, 28.9, 27.4, 25.4, 22.7, 21.4, 20.8, 19.8, 18.4, 16.5, 16.4, 14.8, 14.7 ppm, (ESI) m/z 572.44(M+H)+
(1R,3aR,5aR,5bR,11aR,E)-3a,5a,5b,8,8,11a-hexamethyl-1-(prop-1-en-2-yl)octadecahydro-1H-cyclopenta[a]chrysen-9(5bH)-one O-4-chlorobenzoyl oxime (4b)
Colourless gum, IR (KBr) cm-1 1629 (C=N), 1551 (C=C), 1H NMR (300 MHz, CDCl3): δ = 7.99 (2H, d, J=8.0Hz), 7.43(2H, t, J=8.1Hz), 4.69 (1H, s), 4.57 (1H, s), 2.91 (1H, J = 16.2), 2.55-2.35 (2H, m), 1.90-1.99 (1H, m), 1.91 (1H, q, J = 19.2, 8.8), 1.68 (3H, s), 1.31 (3H, s), 1.19 (3H, s), 1.07 (3H, s), 0.96 (3H, s), 0.94 (3H, s), 0.79 (3H, s), 2.19 (1H, m), 1.56 (4H, m), 1.32 (4H, m), 1.25 (1H, m), 1.67 (1H, m), 1.37 (1H, m), 1.38 (1H, m), 1.45 (4H, m), 1.20 (4H, m), 1.49 (2H, m). 13C NMR (75 MHz, CDCl3): δ = 176.7, 163.4, 150.8, 139.3, 130.8, 128.7, 109.3, 55.3, 49.9, 48.3, 47.9, 42.9, 41.5, 40.8, 39.9, 39.0, 38.0, 37.1, 35.4, 33.6, 29.8, 27.4, 25.0, 22.7, 21.3, 20.0, 19.2, 19.0, 17.9, 16.0, 15.8, 14.4 ppm (ESI) m/z : 578.37 (M+H)+
(1R, 3aR, 5aR, 5bR, 11aR, E)-3a, 5a, 5b, 8, 8, 11a – hexamethyl –1 -(prop –1 –en – 2 -yl) octadecahydro -1H – cyclopenta [a] chrysen-9 (5bH) – one O- piperidine-1- carbonyl oxime (4c)
Pale yellow gum, IR (KBr) cm-1 1639 (C=N), 1549 (C=C), 1H NMR (300 MHz, CDCl3): δ =4.68 (1H, s), 4.57 (1H, s), 3.76 (4H, t, J =9.3, 4.5 Hz) , 3.2 (4H, d, J =2.1,10.5 Hz), 2.59 (4H, t, J =1.2, 1.4Hz), 2.4-2.34 (1H, m), 1.96-1.87 (1H, m), 1.68 (3H, s), 1.25(3H, s), 1.03 (3H, s), 0.94 (3H, s), 0.85 (3H, s), 0.84 (3H, s), 0.78 (3H, s), 1.39 (3H, m),1.32 (4H, m),1.28 (1H, m), 1.68 (1H, m), 1.42 (1H, m), 1.39 (1H, m), 1.48 (4H, m), 1.19 (4H, m), 1.56 (2H, m). 13C NMR (75 MHz, CDCl3): δ = 169.4, 151.0, 109.3, 81.4, 66.8, 59.7, 53.2, 50.2, 48.2, 47.9, 42.9, 42.8, 40.8, 39.9, 38.3, 38.0, 37.8, 35.5, 37.0, 34.1, 29.8, 29.6, 28.0, 27.4, 23.8, 20.9, 18.1, 17.9, 16.5, 16.1, 15.9, 14,4 ppm (ESI) m/z 551.45(M+H)+
Compound 4d
Pale yellow gum, IR (KBr) cm-1 1633 (C=N), 1556 (C=C), 1H NMR (300 MHz, CDCl3): δ = 9.33 (1H, d, J = 1.3 Hz), 8.78 (1H, d, J = 1.3 Hz), 8.74 (1H, t, J = 8.1, 1.7 Hz), 4.68 (1H, s), 4.57 (1H, s), 2.61-2.54 (1H, m), 1.98-1.89 (1H, m), 1.67 (3H, s), 1.34 (3H, s), 1.28 (3H, s), 1.07 (3H, s), 0.96 (3H, s), 0.94 (3H, s), 0.79 (3H, s), 2.16 (1H, m),1.53 (4H, m), 1.29 (4H, m),1.27 (1H, m), 1.67 (1H, m), 1.42 (1H, m), 1.33 (1H, m), 1.49 (4H, m), 1.19 (4H, m), 1.54 (2H, m). 13C NMR (75 MHz, CDCl3): δ = 177.9, 161.7, 150.9, 147.5, 146.1, 144.4, 109.3, 55.2, 49.9, 48.1, 47.9, 42.8, 41.7, 39.9, 39.0, 38.0, 37.1, 35.4, 29.8, 27.3, 26.1, 25.8, 25.0, 22.7, 21.3, 20.1, 19.2, 19.0, 17.9, 6.1, 14.4ppm (ESI) m/z : 546.40(M+H)+
(1R, 3aR, 5aR, 5bR, 11aR, E)-3a, 5a, 5b, 8, 8, 11a-hexamethyl-1- (prop-1-en-2-yl) octadecahydro-1H-cyclopenta [a]chrysen-9 (5bH)-one O-2, 6-dichloro-3-nitrobenzoyl oxime (4e)
Pale yellow gum, IR (KBr) cm-1 1635 (C=N), 1553 (C=C), 1H NMR (300 MHz, CDCl3): δ = 7.39 (1H, d, J = 7.0 Hz), 7.35 (1H, d, J = 7.4 Hz), 4.68 (1H, s), 4.56 (1H, s), 2.88-2.83 (2H, q) 1.67 (3H, s), 1.37 (3H, s), 1.25 (3H, s), 1.06 (3H, s), 0.94 (3H, s), 0.93 (3H, s), 0.79 (3H, s), 2.18 (1H, m), 1.22-1.14 (4H, m), 1.31 (4H, m), 1.28 (1H, m), 1.68 (1H, m), 1.42 (1H, m), 1.39 (1H, m), 1.49 (4H, m), 1.12 (4H, m), 1.43-1.36 (2H, m). 13C NMR (75 MHz, CDCl3): δ = 176.5, 150.8, 147.0, 136.0, 132.7, 132.4, 130.8, 128.7, 127.8, 113.6, 109.3, 68.1, 55.3, 49.9, 48.1, 47.9, 42.8, 41.6, 40.8, 39.9, 39.1, 38.0, 37.1, 35.4, 33.6, 29.7, 27.3, 25.0, 23.7, 22.5, 21.3, 19.8, 18.9, 17.9, 15.8, 14.4 ppm (ESI) m/z 657.31(M+H)+
(1R, 3aR, 5aR, 5bR, 11aR, E)-3a, 5a, 5b, 8, 8, 11a-hexamethyl-1-(prop-1-en-2-yl) octadecahydro-1H-cyclopenta [a] chrysen-9 (5bH)-one O-3-bromo-4-fluoro benzoyl oxime (4f)
Pale yellow gum, IR (KBr) cm-1 1631 (C=N), 1553 (C=C), 1H NMR (300 MHz, CDCl3): δ = 8.25 (1H, dd, J=8.0, 1.9 Hz), 8.01 (1H, dddd, J=2.2, 8.2 Hz), 7.18 (1H, t, J= 8.3, 2.0 Hz), 4.69 (1H, s), 4.57 (1H, s), 2.92-2.87 (1H, q), 1.68 (3H, s), 1.31 (3H, s), 1.19 (3H, s), 1.07 (3H, s), 0.94 (3H, s), 0.96 (3H, s), 0.79 (3H, s), 2.55-2.49 (1H, m), 1.59 (4H, m), 1.34 (4H, m), 1.27 (1H, m), 1.69 (1H, m), 1.43 (1H, m), 1.35 (1H, m), 1.44 (4H, m), 1.25 (4H, m), 1.52 (2H, m). 13C NMR (75 MHz, CDCl3): δ = 177.2, 162.5, 150.8, 149.1, 138.4, 133.1, 128.7,125.5, 109.3, 55.3, 49.9, 48.2, 47.9, 42.8, 40.8, 39.9, 39.0, 38.0, 35.6, 33.7, 30.3, 29.8, 26.8, 22.6, 21.3, 20.0,20.1, 19.3, 17.9, 16.0, 15.8, 14.5ppm (ESI) m/z 640.31(M+H)+
(1R,3aR,5aR,5bR,11aR,E)-3a,5a,5b,8,8,11a-hexamethyl-1-(prop-1-en-2-yl)octadecahydro-1H-cyclopenta[a]chrysen-9(5bH)-one O-3-fluoro-4-methylbenzoyl oxime (4g)
Pale yellow gum, IR (KBr) cm-1 1632 (C=N), 1554 (C=C), 1H NMR (300 MHz, CDCl3): δ = 8.59 (1H, brs), 8.17 (1H, dd, J = 8.2, 1.6 Hz), 7.47 (1H, d, J = 7.9 Hz), 4.68 (1H, s), 4.57 (1H, s), 2.69 (3H, s), 2.57 (1H, m), 1.67 (3H, s), 1.31 (3H, s), 1.21 (3H, s), 1.07 (3H, s), 0.96 (3H, s), 0.93 (3H, s), 0.79 (3H, s), 2.42-2.45 (1H, m), 1.51 (4H, m), 1.37 (4H, m), 1.25 (1H, m), 1.68 (1H, m), 1.45 (1H, m), 1.38 (2H, m), 1.49 (4H, m), 1.19 (4H, m), 1.54 (2H, m). 13C NMR (75 MHz, CDCl3): δ = 175.4, 162.3, 150.8, 149.1, 138.4, 133.4, 133.1, 129.0, 128.1, 125.5, 109.3, 55.3, 48.9, 48.1, 47.9, 42.9, 42.8, 41.6, 39.9, 39.0, 38.0, 33.6, 37.1, 35.4, 33.6, 29.7, 29.2, 26.5, 25.0, 22.7, 21.3, 20.6, 19.2, 19.0, 17.9, 16.0, 15.8, 14.4 ppm (ESI) m/z 576.41(M+H)+
(1R,3aR,5aR,5bR,11aR,E)-3a,5a,5b,8,8,11a-hexamethyl-1-(prop-1-en-2-yl)octadecahydro-1H-cyclopenta[a]chrysen-9(5bH)-one O-2-chloro nicotinoyl oxime (4h)
Pale yellow gum, IR (KBr) cm-1 2995(ArC-H), 1633 (C=N), 1556 (C=C), 1H NMR (300 MHz, CDCl3): δ = 8.53 (1H, dd, J= 7.5, 1.9 Hz ), 8.15 (1H, dd, J= 7.8, 1.9 Hz), 7.35 (1H, t, J= 8.0, 2.2 Hz), 4.68(1H, s), 4.57 (1H, s), 2.56-2.48 (2H, q), 1.68 (3H, s), 1.40 (3H, s), 1.30(3H, s), 1.18 (3H, s), 1.15 (3H, s), 0.93 (3H, s), 0.79 (3H, s), 2.17 (1H, m),1.56 (4H, m),1.31(4H, m),1.26 (1H, m), 1.69 (1H, m), 1.43 (1H,m), 1.38 (1H, m), 1.51 (4H, m), 1.21 (4H, m), 1.53 (2H, m). 13C NMR (75 MHz, CDCl3): δ = 177.3, 163.0, 151.7, 150.8, 140.2, 128.2, 109.3, 55.4, 50.0, 47.9, 48.1, 42.9, 41.7, 40.8, 39.9, 39.2, 37.1, 35.4, 33.7, 29.7, 27.3, 25.0, 22.6, 21.3, 20.4, 19.2, 19.0, 16.1, 15.8, 14.4 ppm (ESI) m/z 579.36(M+H)+
(1R, 3aR, 5aR, 5bR, 11aR, E)-3a, 5a, 5b, 8, 8, 11a-hexamethyl-1- (prop-1-en-2-yl) octadecahydro-1H-cyclopenta[a]chrysen-9(5bH)-one O-2,4-dimethoxybenzoyl oxime (4i)
Colourless amrpous powder,1H NMR (300 MHz, CDCl3): δ = 7.88 (1H, d, J = 7.8 Hz ), 6.53(1H, dd, J = 2.2, 8.6 Hz), 6.51(1H, d, J = 2.2 Hz), 4.68 (1H,s), 4.56 (1H, s), 3.86 (3H, s), 3.85 (3H, s), 2.51-2.5 (2H, q), 1.67 (3H, s), 1.31 (3H, s), 1.18 (3H, s), 1.06 (3H, s), 0.95 (3H, s), 0.94 (3H, s), 0.79 (3H, s), 2.18 (1H, m), 1.55 (4H, m),1.31 (4H, m),1.28 (1H, m), 1.68 (1H, m), 1.42 (1H,m), 1.39 (1H, m), 1.49 (4H, m), 1.19 (4H, m), 1.53 (2H, m). 13C NMR (75 MHz, CDCl3): δ = 177.8, 164.1, 160.8, 150.9, 13.9, 112.0, 109.3, 104.6, 98.9, 55.4, 50.0, 47.9, 42.9, 41.4, 40.8, 39.9, 39.1, 39.1, 38.0, 37.1, 35.5, 33.7, 29.8, 27.3, 22., 25.2, 21.3, 17.9, 16.1, 15.8, 14.4ppm (ESI) m/z: 604.43(M+H)+
(1R, 3aR, 5aR, 5bR, 11aR, E)-3a, 5a, 5b, 8, 8, 11a-hexamethyl-1-(prop-1-en-2-yl) octadecahydro-1H-cyclopenta[a] chrysen-9(5bH)-one O-thiophene-2-carbonyl oxime (4j)
Pale yellow gum, IR (KBr) cm-1 3015(ArC-H), 1633 (C=N), 1556 (C=C), 1H NMR (300 MHz, CDCl3): δ = 7.86 (1H, dd, J = 6.2, 1.2 Hz ), 7.56 (1H, dd, J = 6.2, 1.2 Hz), 7.13 (1H, dd, J = 8.6, 2.2 Hz) 4.68 (1H,s), 4.56 (1H, s), 2.54-2.35 (2H, q), 1.67 (3H, s), 1.30 (3H, s), 1.07 (3H, s), 0.95 (3H, s), 1.15 (3H, s), 0.93 (3H, s), 0.80 (3H, s), 2.16 (1H, m), 1.52 (4H, m), 1.28 (4H, m),1.30 (1H, m), 1.62 (1H, m), 1.42 (1H, m), 1.38 (1H, m), 1.50 (4H, m), 1.20 (4H, m), 1.56 (2H, m). 13C NMR (75 MHz, CDCl3): δ = 176.3, 160.0, 150.9, 132.3, 127.7, 109.3, 56.6, 49.9, 48.2, 48.0, 42.9, 42.8, 41.5, 40.8, 40.0, 39.8, 38.0, 37.1, 35.4, 33.7, 29.8, 27.3, 25.0, 22.7, 19.9, 19.2, 17.9, 16.0, 14.4ppm (ESI) m/z = 550.36 (M+H)
Results and discussion
The synthetic route to oxime ester derivatives conjugated at the C-3 position of Lupeol 1 which was isolated from Walsura trifoliate22 is outlined in Scheme 1. The Lupeol 1 was reacted with PCCin DCM followed by treatment with hydroxyl amine hydrochloridegave the oxime compound 3. The oxime 3 was derivatized with different acids using 2,4,6-trichloro benzoyl chloride, triethyl amine, DMAP under yamaguchi etherification conditions to afford different oxime ester derivatives (4a-4j) showed in table -1 .
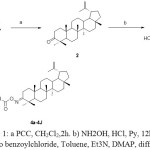 |
Scheme 1: a) PCC, CH2Cl2,2h. b) NH2OH, HCl, Py, 12hr, c) 2,4,6 tri chloro benzoylchloride, Toluene, Et3N, DMAP, different acids. Click here to View scheme |
 |
Table 1: Different oxime ester derivatives, time and yield (4a-4j) Click here to View table |
Table2 : Antibacterial activates of lupeol derivatives (1-4J) MIC (µg/ml)
| Compound code | B.Subtilis | S.aureus | S.epidermidis | E.coli | P.aeroginosa | K.pneumoniae |
| 1 | 100 | 150 | 75 | 150 | 150 | 150 |
| 2 | 75 | 100 | 50 | 100 | 75 | 50 |
| 3 | 25 | 75 | 75 | 150 | 150 | 150 |
| 4a | 150 | 100 | 100 | 150 | 150 | 150 |
| 4b | 50 | 75 | 50 | 150 | 110 | 100 |
| 4c | 100 | 100 | 100 | 150 | 150 | 150 |
| 4d | 75 | 150 | 150 | 100 | 75 | 150 |
| 4e | 100 | 100 | 75 | 100 | 150 | 75 |
| 4f | 150 | 100 | 100 | 75 | 150 | 150 |
| 4g | 100 | 150 | 100 | 150 | 150 | 150 |
| 4h | 75 | 50 | 50 | 25 | 100 | 50 |
| 4i | 100 | 150 | 150 | 150 | 150 | 150 |
| 4j | 150 | 75 | 100 | 100 | 75 | 50 |
| Penicillin | 1.562 | 1.562 | 3.125 | 12.5 | 12.5 | 6.25 |
| Streptomycin | 6.25 | 6.25 | 3.125 | 6.25 | 1.562 | 3.12 |
Antibacterial activity of Lupeol and its derivatives
All the lupeol and its derivatives were tested against antimicrobial activity using standard protocol24 against three gram positive bacteria viz. Bacillus subtilis (MTCC-4411), Staphylococcus aureus (MTCC-96), Staphylococcus epidermidis (MTCC-2639) and three gram-negative bacteria viz. Escherichia coli (MTCC-443), Pseudomonas aeruginosa (MTCC-741), and Klebsiella pneumoniae (MTCC-618). The MIC of the compounds was tabulated in table-2. Standard drugs like Penicillin and Streptomycin were taken for comparison. Lupeol and its derivatives are inactive activity against all the bacterial strains showing high values.
Anti-proliferative activity of Lupeol and its derivatives
The invitro anti-proliferative activity of isolated lupeol and its derivatives were examined against the cell lines- MDAMB-231(breast cancer), IMR32 (neuroblastoma) and A549 (lung cancer) following standard protocol23. The reference standard used for the antiproliferative activity was Doxorubicin which was expressed as GI50 values (Growth inhibition 50 % μg/mL) the results were tabulated in table-3. The results revealed that some of the synthetic analogues were exhibited promising anticancer activity when compared their parent isolated compound. Among the tested compounds, compound 3 showed good activity 20.8±0.09 (IMR 32). While compounds 4e, 4f &4h showed moderate activities with GI50 values of 28.7±0.2, 22.4±0.09 & 27.5±0.4 /mL respectively. It is important to mention that all the tested compounds were not active against IMR 32, MDAMB-231, and A549 Cell lines. Different substitutions on the aromatic ring, derivatives affect the activity. Though it is difficult to discuss the structure activity relationship criteria responsible for anti-proliferative activity in this set of compounds
Table 3: Anti-proliferative activity of lupeol(1) derivatives (GI50 in µg/mL)
| sample | MDA-MB-231 | IMR 32 | A549 |
| 1 | – | >100 | >100 |
| 2 | – | 33.7±0.1 | 23.5±0.4 |
| 3 | – | 20.8±0.09 | – |
| 4a | – | – | – |
| 4b | – | – | 31.5±0.1 |
| 4c | – | – | – |
| 4d | – | 32.6±0.2 | – |
| 4e | 28.7±0.2 | – | – |
| 4f | – | 22.4±0.09 | – |
| 4g | 32.7±0.08 | 51.0±0.7 | 52.6±0.4 |
| 4h | – | 27.5±0.4 | 28.3±0.04 |
| 4i | – | – | – |
| 4j | 96.4±0.3 |
85.6±0.2 |
91.2±0.5 |
Conclusion
In conclusion, synthesis of series of oxime ester derivatives from natural product Lupeol has been achieved using PCC oxidation, oxime formation followed by yamaguchi etherification. These Synthesized derivatives can be screened for their antimicrobial and anti-proliferative activity studies. All the compounds showed the modatare actives.
Acknowledgements
The authors like to thank for the support of Divis laboratories for the support given for completion of the work
References
- Connolly, J. D.; Hill, R. A. Nat. Prod. Rep. 1997, 14, 661–679.
CrossRef - Hill, R. A.; Connolly, J. D. Nat. Prod. Rep. 2012, 29, 780–818
CrossRef - Suri Appa Rao, M.; Suresh, G.; Ashok Yadav, P.; Rajendra Prasad, K.; Usha Rani, P.; Venkata Rao, C.; Suresh Babu, K. Tetrahedron. 2015, 71, 1431-1437
CrossRef - Murthy, K. S. R.; Nagamani, K. J. J. Toxicol. 2008, 3, 267-271
- Anjaneyulu, V.; Prasad, K.H.; Rao, G.S. Indian J.Pharm. Sci. 1982, 44, 58–59
- Geetha, T.; Varalakshmi, P. J. Ethnopharmacol. 2001, 76, 77-80
CrossRef - Geetha, T.; Varalakshmi, P. Mol Cell Biochem. 1999, 201, 83-87
CrossRef - Sunitha, S.; Nagaraj, M.; Varalakshmi, P. Fitoterapia. 2001, 72, 516-23
CrossRef - Nagaraj, M.; Sunitha, S.; Varalakshmi, P. J. Appl.Toxicol. 2001, 20, 413–417.
CrossRef - Ulubelen, A.; Topcu, G.; Johansson, C. B. J Nat Prod. 1997, 60, 1275-80.
CrossRef - Fátima Gutiérrez-Nicolás.; Bárbara Gordillo-Román.; Juan, C.; Oberti.; Ana Estévez-Braun.; Ángel, G.; Ravelo.; Pedro Joseph-Nathan. J. Nat. Prod. 2012, 75, 669–676.
- Chaturvedi, P. K.; Bhui, K.; Shukla, Y. Cancer Lett. 2008, 263, 1-13.
- CrossRef
- Murtaza, I.; Saleem, M.; Adhami, V. M.; Hafeez, B. B.; Mukhtar, H. Cancer Res. 2009, 69,1156-65
CrossRef - Mohammad Saleem.; Farrukh Afaq.; Vaqar Mustafa Adhami.; Hasan Mukhtar. Oncogene. 2004, 23, 5203–5214
CrossRef - Sudharsan, P. T.; Mythili, Y.; Sudhahar, V.; Varalakshmi, P. J Pharm Pharmacol. 2005 57, 1437-44
CrossRef - Sudharsan, P. T.; Mythili, Y.; Selvakumar, E.; Varalakshmi, P. Mol Cell Biochem. 2006, 292, 39-44
CrossRef - Vijai Lakshmi.; Abbas Ali Mahdi.; Mohammad Kaleem Ahmad.; Santosh Kumar Agarwal.; Arvind Kumar Srivastava. Bangladesh Pharmaceutical Journal. 2014, 17, 138-146.
- Varatharajan Sudhahar.; Sekar Ashok Kumar.; Yenjerla Mythili.; Palaninathan Varalakshmi. Nutrition Research. 2007, 27, 778–787
CrossRef - Varatharajan Sudhahar.; Sekar Ashok Kumar.; Periyasamy Thandavan Sudharsan.; Palaninathan Varalakshmi. Vascular Pharmacology. 2007, 46, 412–418.
CrossRef - Linday, M. E. Practical Introduction to Microbiology; E and F. N. Spon ltd: United Kingdom, 1962.
- National Committee for Clinical Laboratory (NCCL). 5th ed.; Approved Standard M7-A5, NCCLS: Villanova, PA, 2000.
- Murthy, K. S. R.; Nagamani, K. J. J. Toxicol. 2008, 3, 267-271
- Pavan Kumar, Ch.; Devi, A.; Ashok Yadav, P.; Rao Vadaparthi, R.; Shankaraiah, G.; Sowjanya, P.; Nishant Jain.; Suresh Babu, K. Journal of Asian Natural Products Research, 2016,11, 1063-1078
CrossRef - Reddy.; Pedavenkatagari Narayana., Pannala Padmaja.; Bobbala Ramana Reddy.; Gundla Rambabu.; Machiraju Pavan Kumar. Medicinal Chemistry Research, 2016, 1-11

This work is licensed under a Creative Commons Attribution 4.0 International License.









