Cholesterol Interactions with Fatty Acids and DMPC Phospholipids of Liver Membranes
Arina Rahimi and Majid Monajjemi*
Department of Chemistry, Science and research Branch, Islamic Azad University, Tehran, Iran.
Corresponding Author E-mail: m_monajjemi@srbiau.ac.ir
DOI : http://dx.doi.org/10.13005/ojc/320615
Cholesterol and fatty acidsis important subject in liver to different model of regulation for realizing the evolution of vertebrates. The major solubility of cholesterol in bilayers of glycerol-phospholipids is between 65 and 50 mole%,relevant on the bilayerof lipid membrane but they cannot alone form multi layered structures. Livers from the transgenic rat showed increases in mRNAs encoding various enzymes of cholesterol synthesis, the LDL’s receptor and fatty acid synthesis. Based on our previous works we have modeled and simulated various molecules of that Cholesterol in binding to membrane. A number of computational chemistry studies carried out to understand of the cholesterol parallel to fatty acid synthesis (FAS) for preventing the fatty liver disease.In this work ELF, LOL, ECP, electrical properties such as electron densities, energy densities, and potential energy densities, eta index forsome of the fatty acidshave been calculated.
KEYWORDS:Cholesterol; Fatty acid; liver; Nano-biotechnology; drug design
Download this article as:| Copy the following to cite this article: Rahimi A, Monajjemi M. Cholesterol Interactions with Fatty Acids and DMPC Phospholipids of Livermembranes. Orient J Chem 2016;32(6). |
| Copy the following to cite this URL: Rahimi A, Monajjemi M. Cholesterol Interactions with Fatty Acids and DMPC Phospholipids of Livermembranes. Orient J Chem 2016;32(6). Available from: http://www.orientjchem.org/?p=25288 |
Introduction
In contrast to the diversity of phospholipid types, the cells of mammals contain one main sterol, cholesterol, that is exactly required forlivability and cell proliferation1, 2. Sterols vary from the other membrane bilayer classes, and originally consist of hydrocarbon in the form of a steroid structures. The maximum solubility of cholesterol in bilayers of membranes is between 65 and 50 mole%,relevant on the bilayerof lipid membrane3,4but they cannot alone form multi layered structures5. Sterols become manifest to havesolved for filling the spaces amongst the acyl chains in the membranes6
Fatty acid, cholesterol and the primitive lipids synthesized in the liver are a subject to different template of regulation and synthesis. Cholesterol synthesis is great when animals are fed a free diet cholesterol, and it deteriorationsignificantlyduring of cholesterol feeding. Cholesterol synthesis is also increase when animals consume bile acid–binding resins.
In biological membrane phospholipid bilayers, the length and impregnation of the phospholipids and hydrocarbon also chip in to the different properties of sphingomyelin and phosphatidylcholine’s. Generallyhappeningphosphatidylcholine’susuallyare including two hydrocarbon of approximately similar length.
The attached “acyl” chain to the first carbon of glycerol is generally saturated, while the “acyl” chain esterified to the C2 (next carbon) is normally unsaturated3-5.For more understanding of “acyl” chain whichbasically containsbetween 1 and 6 double bonds of cis-configuration we simulated our systems. The sphingomyelins in membranes are extremely saturated than naturally phosphatidyl-cholines6. In addition, natural sphingomyelinalways has very long amide-linked “acyl” chain carbon in the length, while the sphingoid base is most often sphingosine (d-erythro-2amino-trans4-octadecene-1, 3diol).
This will lead to an inconformity in chain length for more natural-sphingomyelins, and such a chain difference has been shown to improve membrane curvature7, 8.
The chain disparities also enable sphingomyelins to participate in the trans-bilayer hydrocarbon inter-digitation.
The molecular structure of the cholesterols includes a skeleton ( tetracyclic fused ring skeleton), with a alone hydroxyl group at carbon three, a double bond between two carbons “five and six” and an isooctyl hydrocarbons side chain at the carbon seventeen.
In Fig. 1 cholesterol bound from human beta2 adrenergic receptor has been shown9, 10. The rings of the cholesterol ismolten in the trans configuration, which makes the moleculerigid and planar, except for the isooctyl side chain11-13..An important concept on the three dimensional structure of the cholesterol is that the “3b-OH” group, 2 methyl groups and the side chains are all located on the same side of the ring skeleton4,15.
The hydroxyl group in the cholesterol is important, because it gives another hydrophobic compound (its amphiphilic character) and therefore orients the molecule in phospholipids membrane.
Further, the “OH” group can also interfere the hydrogen bonding of cholesterol byH2O and possibly with other lipids of cellular membranes16, 17.
Another interesting consequence of the cleavage of lipids bilayers by phospholipases is that the lateral organization of membrane bilayers may be drastically altered during the enzymatic activity. It has been confirmed where the characterizing effects of cholesterol on the line tension of solid DMPC domains vanishes rapidly after the phospholipase activity sets in. This is an instance of the tight coupling betweenenzyme activity and membrane bilayers structure alterations.People are worried about eating cholesterol due to a relationship to coronary heart disease.
Dietary restrictions of cholesterol intact do notsolve this problem, since we make most of the
cholesterol in our bodies (~70% on average). We need cholesterol: it maintains membrane
fluidity and forms the basic skeleton for steroidsand hormones. From the looks of it, you may think (it seems) that this mustcome from a lot of different carbon skeletons. The Biosynthesis of Cholesterol has been drawn in Fig.2
It is most important for understanding the natural &pharmaceutical-based Regulation of Sterol / Cholesterol. Production is based on cholesterol synthesis is an energy-expensive complex process in the cell membrane. Thus it is heavilyregulated. The major committed, rate-limiting step of sterol production is the formation of Mevalonate from βHydroxy-βmethylglutarylCoA by the enzyme “HMGCoA”reductase. Therefore, popular target of manydrugs, called statins.
Statins and all derivatives of fungalnatural products Fig3.
Livers from the transgenic rat showed increases in mRNAs encoding various enzymes of cholesterol synthesis, the LDL’s receptor and fatty acid synthesis. The elevations in mRNA for 3hydroxy-3methylglutaryl coenzyme “A (HMG CoA)” synthase and “HMG CoA”reductase were especially marked.
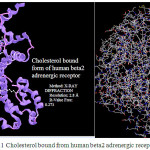 |
Figure 1: Cholesterol bound from human beta2 adrenergic receptor |
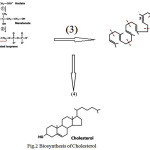 |
Figure 2: Biosynthesis of Cholesterol |
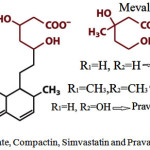 |
Figure 3: Mevalonate, Compactin, Simvastatin and Pravastatin structures |
Theoretical Background
Density and Energy
The density of electron can be defined as
![]()
Where ηi is “occupation” number of orbital (i), is wave function orbital, χ is the basis function and C is
the coefficient matrix, the element of row “ith” and column “jth” corresponds to the expansion coefficient
of orbital “j”respect to basis function “i”. Atomic unit for density of electron can be explicitly written as
e/Bohr3.
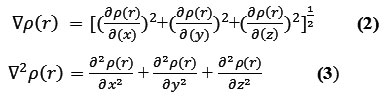
The positive and negative value of this function correspond to electron density is locally concentrated and depleted respectively. The relationships between ▽2ρ and VSEPR (valence shell electron pair repulsion) model, electron localization,chemical bond type and chemical reactivity have been investigated by Bader .
The kinetic energy density is not individually defined, since the expected amount of kinetic energy operator
![]()
must be recovered by integrating of kinetic energy density from the alternative definitions.
One of important used definition is
![]()
Relative to K(r), the local kinetic energy definition given below guarantee positivizes everywhere; hence the physical meaning is clearer and is more commonly used.
The G(r) “Lagrangian” of kinetic energy density is also known as the positive definite energy density.
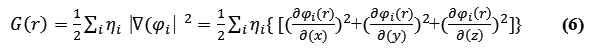
K(r) and G(r) are straightly defined by Laplacian of electron density
![]()
Edgecombe and Becke, noted that averaged like spin conditional (pair probability) has direct relation with the “Fermi hole” and then suggested ELF (electron localization function)18-20.
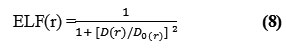
where,
![]()
and
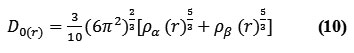
for close-shell system, since
![]()
D and D0 terms can be simplified as
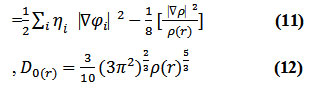
Savinhas reinterpreted “ELF” in the viewpoint of kinetic energy20 which makes “ELF” also meaningful for “Kohn-Sham” DFT wave-function or even post-HF wave-function20. They indicated which“D(r)” reveals the excess kinetic density caused through Pauli repulsion, while D0(r) can be evaluated as “Thomas-Fermi” kinetic energy density19,20.
LOL or Localized orbital locator is the other function for locating high localization regions as the same “ELF”, defined by Becke and Schmider.
![]()
where
![]()
Computational Details
Calculations were accomplished by Gaussian package. In present work, we have looked atfor getting the results from “DFT” methods such as m062x, m06L, and m06 for the molecular reaction that are monotonous via the comparison between different situations.
The m062x, m06L, and “m06HF” are new approach in DFT with a good correspondence in reaction calculation and are useful for the energy between two fragments in cholesterol with binding to a membrane.
We employed DFT theory to model the exchangecorrelation energies for our systems. The double ζ-basis sets with a polarization orbital (DZP) were used for the phospholipids atoms while single ζ-basis set with a polarization orbital (SZP) were employed for the fatty acids in the membrane layers, respectively.
For an accurate DFT calculation; “B3LYP” is not able to describe correlationsystems by interaction. The disability of B3LYP and other popular functional to correctly describe of correlation and exchange limits their applicability for the two parts. Recently, studies have shown that inexactitude for the exchange energy leads to the large systematic errorfor the prediction of chemical physics properties for the molecules.
Electronic structure calculations and geometry optimizationfor our systems have been performed using the “Kohn-Sham” equation of the DFT theory for a plane wave basis set with the projectoraugmented wave pseudopotentials.
The PBU (Perdew-Burke-Ernzerh) of XC(exchange-correlation) functional of the GGA(generalized gradient approximation)is adopted. The optimization of the lattice constant and the atomic coordinate are made by the minimization of the total energy. Except for the band determination, the partial occupancy was treated using the tetrahedron methodology with Blöchlcorrection.
The charge transfer and electrostatic potential-derived charge was also calculated using the Merz-Kollman-Singh, chelp, or chelpG.
For the charge calculation, based on molecular MESP (electrostatic potential)fitting are not well suited for treating a large system where some of the atoms are located far away from the points at which the “MESP” is computed. In this condition, variation of those atomic charges will not lead to significant changes of the outset atoms for the molecule. In other words, the accurate values for the innermost atomic charges are not well-determined via MESP of the molecule.
The representative atomic charge for our molecules should be computed as average value over several phospholipid molecules. We have also extracted the charge densities profiles from firstprinciples calculationsvia an averaging process described. Based on our previous works21-46 the system of cholesterol in binding to DMPC has been simulated. The result has shown that the mechanism of fatty acid synthesis (FAS)enhanced due to cholesterol increases extremely.
Result and Discussion
Monte Carlo simulation always detect the socalled “important phase-space” regions which are of a low energy. Because of defect of the force field, those lowest energies basin usually does not correspond to a native state in most cases. So the rank of the native structures in those decoys has produced through the force fields it selvesare poor. In this study difference in force field is illustrated by comparing the energies and physical properties calculated by using molecular mechanic force fields such as MM+, Amber, and OPLS.
This study has investigated mainly on the electron density of cholesterol in a binding system with DMPC membrane. The binding details are shown in figs1-6. As it has been indicated in table1, the electrical and magnetically properties can be obtained via changes in the binding interaction.
Electron densities such as energy density, Potential energy density,eta index, “ELF”, “LOL”, electron density, and ECP forcholesterol were calculated for each simulation.
According to the equation 10- 14 the largestELF is located onatoms which are bonded to phosphor of phospholipids which in those atomsthe electron motion is more likely to be confined within that region. If an electron is completely localized in such atoms, they can be distinguished from the other ones. As it has been shown in the tables2&3 the largest ELF is close to the bonded atoms to phosphors. The regions with large ELFneed to have large magnitudes of Fermi-hole integration which would leadthose atoms towards “superparamagnetic” phenomenon.The “Fermi hole” is a six-dimension function and as a result, it is difficult to be studied visually12-14.
Based on equations 10-14, Edgecombeand Becke have noted that the “Fermi hole” is a spherical average of spin which is in a good agreementwith our results.
“ELF”indicates that it is a relativelocalization and must be accounted within the range of [0-1]. A large amount of ELF value corresponds to a large localized electronwhich indicates that a lone pair or inner shells of the atom are involved12-14. According to equation 14,“LOL” can be interpreted similar to the ELF in terms of a kinetic energy,though; “LOL” can also be interpreted in terms of localized orbitals. Large (small) LOL value usually appears in the boundary (inner) region of the localized orbitals due to the small (large) gradient of orbital wavefunction in this area. The value range of “LOL” is identical to “ELF”, namely [0, 1].
 |
Figure 4: Contour line map for Human adipocyte fatty acid |
 |
Figure 5: Cholesterol among 60 domain of DMPC membrane in leaver |
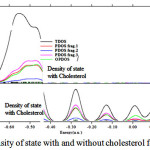 |
Figure 6: Density of state with and without cholesterol for DMPC |
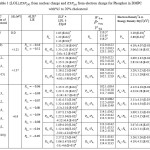 |
Table 1:(LOL), from nuclear charge and from electron charge for Phosphor in DMPC with%5 to 20% cholesterol |
References
- Dahl, C.; Dahl, J.; Boca Raton (FL, USA)CRC Press, 1988, 1, 147–172.
- Yeagle, PL. Boca Raton (FL, USA): CRC Press, 1993. 1–12.
- Israelachvili, JN. Intermolecular and surface forces: with applications to colloidal and biological systems.London (UK): Academic Press, 1989.
- Huang, J.; Buboltz, JT.; Feigenson, GW. BiochimicaetBiophysicaActa1999, 1417(1):89–100.
- Ohvo-Rekila, H et al. / Progress in Lipid Research, 2002, 41, 66–97
CrossRef - Mouritsen, OG. Jorgensen, K. ChemPhys Lipids, 1994, 73(1–2):3–25
CrossRef - Ramstedt, B.; Leppimaki, P.; Axberg, M.; Slotte, JP.;Eur, J Biochem1999;266(3):997–1002.
- Yechiel, E.; Barenholz. Y.; Henis, YL.;J BiolChem1985;260:9132–6.
CrossRef - Urich, K.; In: Comparative animal biochemistry. Berlin: Springer-Verlag, 1994. 624–656.
CrossRef - Edwards, PA.; Davis, R.; In: Vance DE, Vance JE, editors. Elsevier Science, 1996. 341–362.
- Nes WR. Lipids 1973;9:596–612.
CrossRef - Bloch, KE. CRC Crit Rev Biochem1983;14:47–92.
CrossRef - Yeagle, PL.; BiochimBiophysActa1985, 822(3-4):267–87.
CrossRef - Duax ,WL.; Wawrzak, Z.;Griffin, JF.; Cheer, C.;Yeagle, PL.;. Biology of cholesterol.Boca Raton (FL,USA): CRC Press, 1988. 1–18.
- Bittman, R.; In: Bittman, R, editor. Cholesterol: its functions and metabolism in biology and medicine. New York (USA): Plenum Press, 1997. 145-172.
CrossRef - Boggs, JM.; BiochimBiophysActa1987906(3):353–404.
- Bittman, R.; Kasireddy, CR.; Mattjus, P.; Slotte, JP. Biochemistry1994;33(39):11776–81
CrossRef - Lu, T.; Chen, F.;ActaChim. Sinica, 2011, 69, 2393-2406
- Lu, T.; Chen, F.J. Mol. Graph. Model,2012, (38): 314-323
CrossRef - Lu T,; Chen, F.;Multiwfn: A Multifunctional Wavefunction Analyzer, J. Comp. Chem.2012, (33) 580-592
- Monajjemi, M.; Lee, V.S.; Khaleghian, M.; B. Honarparvar, B.; F. Mollaamin, F. J. Phys.Chem C. 2010, 114, 15315
CrossRef - Monajjemi, M.Struct Chem.2012, 23,551–580
CrossRef - Monajjemi, M.; Boggs, J.E. J. Phys. Chem. A, 2013,117,1670 −1684
CrossRef - Mollaamin, F.; Monajjemi, M, Journal of Computational and Theoretical Nanoscience. 2012, 9 (4) 597-601
CrossRef - Monajjemi, M.; Khaleghian, M, Journal of Cluster Science. 2011, 22(4),673-692 31
- Monajjemi, M. Chemical Physics. 2013, 425, 29-45
CrossRef - Monajjemi, M.; Wayne Jr, Robert. Boggs, J.E. Chemical Physics.2014, 433, 1-11
- Monajjemi, M.; Mollaamin, F. Journal of Computational and Theoretical Nanoscience, 2012, 9(12) 2208-2214
CrossRef - Monajjemi, M. Falahati, M.; Mollaamin, F.; Ionics, 2013, 19, 155–164
CrossRef - Monajjemi, M.; Mollaamin, F. Journal of Cluster Science, 2012, 23(2), 259-272
CrossRef - Tahan, A.; Monajjemi, M. Acta Biotheor,2011, 59, 291–312
CrossRef - Lee, V.S.; Nimmanpipug, P.; Mollaamin, F.; Kungwan, N.; Thanasanvorakun, S.; Monajjemi, M. Russian Journal of Physical Chemistry A, 2009,83, 13, 2288–2296
- Mollaamin, F.; Monajjemi, M.Physics and Chemistry of Liquids .2012, 50, 5, 2012, 596–604
- Monajjemi, M.; Khosravi, M.; Honarparvar, B.; Mollaamin, F.; International Journal of Quantum Chemistry, 2011, 111, 2771–2777
CrossRef - Monajjemi, M., Mahdavian, L., Mollaamin, F. Bull. Chem. Soc. Ethiop. 2008, 22(2), 277-286
CrossRef - Monajjemi, M.; Farahani, N.; Mollaamin, F. Physics and Chemistry of Liquids, 2012, 50(2) 161–172
CrossRef - Monajjemi, M. TheorChemAcc, 2015, 134:77 DOI 10.1007/s00214-015-1668-9
CrossRef - Monajjemi, M. Journal of Molecular Modeling, 2014, 20, 2507
CrossRef - Monajjemi, M.; Khaleghian, M.; Mollaamin, F. Molecular Simulation. 2010, 36, 11, 865
- Ilkhani, Ali R.; Monajjemi, M. Computational and Theoretical Chemistry.20151074, 19–25
- Monajjemi, M. Biophysical Chemistry.2015207,114 –127
- Jalilian,H.; Monajjemi, M. Japanese Journal of Applied Physics. 2015,54, 8, 08510
- Mahdavian, L.; Monajjemi, M. Microelectronics Journal. 2010,41(2-3), 142-149
CrossRef - Monajjemi,M *.; Bagheri,S.; Moosavi,M.S..; Moradiyeh,N.; Zakeri,M.; Attarikhasraghi,N.; Saghayimarouf,N.; Niyatzadeh,G.; Shekarkhand,M.; Mohammad S. Khalilimofrad, Ahmadin,H.; Ahadi,M.; Molecules 2015, 20, 21636–21657;
CrossRef - Monajjemi,M.; Nayyer T. MohammadianJ. Comput. Theor.Nanosci.2015,12, 4895-4914.
- Monajjemi, M., Chahkandi, B. Journal of Molecular Structure: THEOCHEM, 2005, 714 (1), 28, 43-60.
CrossRef - Naghsh,F, orient.j.chem2015, 31(1) 465-478
- Chitsazan, A, orient.j.chem, 2015, 31(1) 393- 40
- Barmaki, Z, orient.j.chem2015, 31(3) 1723-1733
- Bonsakhteh, B.; Rustaiyan, A.H, orient.j. chem,2014, 30(4) 1703-1718

This work is licensed under a Creative Commons Attribution 4.0 International License.









