Synthesis, Evaluation and Docking studies of Cholecalciferol Derivative
Sultanat 1,*, Ayaz Mahmood Dar1, Asim Rizvi2, Imrana Naseem2
1Department of chemistry, Aligarh Muslim University, Aligarh 202002 India 2Department of Biochemistry, Aligarh Muslim University, Aligarh 202002 India
DOI : http://dx.doi.org/10.13005/ojc/300322
Article Received on :
Article Accepted on :
Article Published : 23 Aug 2014
Improved synthesis of 3b-acetoxy-9, 10-seco-19, 8(8)-spiro-5(10), 6-cholestadiene has been reported after reacting cholecalciferol acetate with dimethylbutadiene by incorporating BF3·OEt2, SnCl4, ZnBr2, p-TsOH in toluene under Diels-Alder condition to get better yields. Agarose gel electrophoresis showed the potential in vitro DNA damaging nature while as the comet assay depicted the genotoxic nature by mobilizing the tail of the comet in lymphocytes. The molecular docking depicted the intercalation of steroid derivative with minor groove of the DNA molecule and in this configuration the phosphodiester bond of DNA stabilizes the acetoxy group. The bioactivity score and PASS software analysis confirmed the potential physicochemical features of the compound to act as active drug.
KEYWORDS:cholestadiene; cholecalciferol; comet assay; gel electrophoresis; docking
Download this article as:| Copy the following to cite this article: Sultanat, Dar A. M, Rizvi A, Naseem I. Synthesis, Evaluation and Docking studies of Cholecalciferol Derivative. Orient J Chem 2014;30(3). |
| Copy the following to cite this URL: Sultanat, Dar A. M, Rizvi A, Naseem I. Synthesis, Evaluation and Docking studies of Cholecalciferol Derivative. Orient J Chem 2014;30(3). Available from: http://www.orientjchem.org/?p=4328 |
INTRODUCTION
Developments of efficient methodologies to access large molecules are of special interest. Steroidal compounds are essential to life in various ways. Synthetic derivatives of steroids have also attracted a good deal of attention for the purpose of developing lead compounds in order to cure different biological disabilities [1]. Especially synthesizing the steroids comprising heterocycles have received much attention of the chemists since many of such compounds have been shown to display important pharmacological properties [1].
Synthesis of polycyclic skeletons like steroids, have a central place in the development of regio- and stereo chemically controlled synthetic methodology [2]. Boron trifluoride etherate (BF3·OEt2), tin chloride (SnCl4), zinc bromide (ZnBr2), p-toluene sulphonic acid (p-TsOH) has been studied for those reactions which involve formation of carbon-carbon or carbon-heteroatom bonds. Because of various advantages incorporated with these compounds, some studies revealed their uses as potential reagents for numerous reactions involving unexpected formation of dioxaspiro compounds[3]. Some representative examples include cycloaddition, isomerisation, ring contraction, ring expansion and unexpected rearrangement reactions [4-13]. The Diels-Alder pathway is the most studied reaction, showing numerous uses in synthetic procedures and has been intensively studied [14]. The important tool in Diels-Alder chemistry is the activation of dienophiles by Lewis acid [15]. The barrier lies in searching those conditions which promote cycloaddition but without subsequent aromatization. In continuation of our interest in modified steroids [16], we herein report the synthesis of desired cholecalciferol derivative in presence of different non-chelating Lewis acids like BF3·OEt2, SnCl4, ZnBr2, p-TsOH under Diels-Alder condition and simultaneously study its binding with DNA by gel electrophoresis and molecular docking, and genotoxicity by comet assay.
RESULTS AND DISCUSSION
Chemistry
In synthetic organic chemistry, the Diels-Alder reaction is the convenient procedure for forming the 6-membered ring systems with potential control over regio- and stereo chemical properties. This high regio- and stereo selectivity typically have resulted in various uses of this transformation in the building of highly sneaker targets [17]. Diels-Alder procedure has shown mature development, depicting the basic importance in synthetic and theoretical sciences. Now a day, this wonderful procedure is one of the most studied and well reviewed, having a beat of incorporations in synthesis. Formulating hetero complexes with different functionalities and their derivatization is the subject of high concern to synthetic scientists. Here we have investigated cyclization behaviour of cholecalciferol acetate with 2, 3-dimethylbutadiene in presence of different reagents like BF3·OEt2, SnCl4, ZnBr2, p-TsOH under Diels-Alder condition in toluene and different yields were obtained. The synthetic strategy adopted to obtain the target product 2 is revealed in Scheme 1. The product obtained was characterized by IR, 1H NMR and 13C NMR. The characterization showed close agreement with given structure of compound 2. It is observed that the absence of signal at δ 4.2 in the products of 1H NMR is strongly indicating that the exocyclic methylene is involved in cyclization by intramolecular Diels-Alder reaction. Dimethylbutadiene did not played an important role as the same product was obtained in presence and absence of dimethylbutadiene indicating that reagent is not incorporated. Further intramolecular Diels-Alder reaction is occurring involving terminal methylene for cyclization.
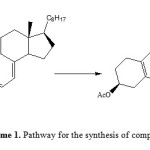 |
Scheme1: Pathway for the synthesis of compound 2 Click here to View Scheme |
It was found that after subjugating different Lewis and Brønsted acids like BF3·OEt2, SnCl4, ZnBr2, p-TsOH and it was found that the maximum yield was obtained in the order BF3·OEt2 > SnCl4 > p-TsOH > ZnBr2.
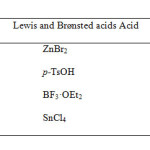 |
Table Click here to View table |
IR spectrum exhibited absorption band at 1026 cm-1 for C-O,1735 cm-1 for carbonyl group of acetate and other band at1655 cm-1 for (-C=C-C=C-). The 1H NMR spectrum displayed a multiplet centred at δ 5.2 for one proton can be ascribed to C3–αH, axial. A distorted signal at δ 5.8- 6.8 can be assigned to the two vinylic protons at C6 and C7. 13C NMR depicted signals at δ 36.3 for C3, δ 138.2 for C5, δ 126.2 for C6, δ 128.2 for C9. The strategy can also be applied to diverse steroidal derivatives.
Evaluation of In vitro DNA damage using plasmid PBR322
In vitro DNA damage of compound 2 was assessed using agarose gel electrophoresis as shown in Figure 1. Lane 1 shows control plasmid, lane 2 depicts plasmid with incubation of 100 µM drug and lane 3 reveals plasmid incubated plus 200 µM drug. It is evident from the disappearance of the band and lytic products observed in the gel that the drug is capable of causing DNA damage. At higher concentration, the disappearance in the band intensity might be the asset of acting the target steroid closely with duplex moiety, which makes the Ethidium bromide to replace.
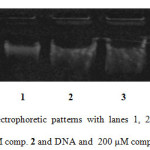 |
Figure1: Agarose gel electrophoretic patterns with lanes 1, 2 and 3 depict DNA only (control), DNA and 100 μM comp. 2 and DNA and 200 μM comp. 2, respectively. Click here to View Figure |
Evaluation of Ex vivo DNA damage using human lymphocytes
Using human lymphocytes, we assessed DNA damage caused by the compound 2 on human genomic DNA ex vivo by comet assay. Figure 2a shows control lymphocyte in which no comet pattern was observed, figure 2b shows lymphocyte incubated with 100 µM drug, figure 2c shows lymphocyte incubated with 200 µM drug and figure 2d shows lymphocyte incubated with 200 µM 5-fluorouracil. The compound 2 did not exhibited apoptotic damage to the level of 5-fluorouracil. Tail length migration presented in (Table 1) show that the molecule is capable of causing severe damage to human genomic DNA. The enhancement in DNA damage suggested that steroid derivative induced dose-dependent chromosomal fragmentation leading to apoptosis. The slides were screened for measuring the length of tail shown by the comet after acting with steroid and the outcome is shown in Figure 3.
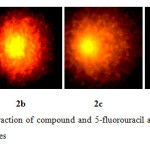 |
Figure2: Depict the interaction of compound and 5-fluorouracil at the various concentrations on the DNA of lymphocytes Click here to View Figure |
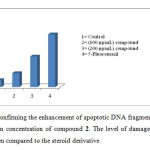 |
Figure3: Graph confirming the enhancement of apoptotic DNA fragmentation in lymphocytes with the increase in concentration of compound 2. The level of damage caused by 5-Fu (100 μM) was more when compared to the steroid derivative.Click here to View Figure |
| Tail Length (µm) | |
| Control lymphocyte | 5 + 0.25 |
| Lymphocyte + 100 µM compound 2 | 10 + 0.36 |
| Lymphocyte + 200 µM compound 2 | 22 + 0.54 |
| Lymphocyte + 200 µM 5-fluorouracil | 38 + 0.41 |
Table 1. Tail length Migration in µm
DNA- compound interaction by molecular docking method
Here the DNA nitrogen base sequence (taken from Protein Data Bank ID: 1BNA) was docked with compound 2 so as to clarify the target point as well as the expected stable interaction of the steroid in the DNA groove. As evident from figure 4 that compound 2 interacts in the minor groove and in this configuration the 3b-acetoxy group intercalates with DNA via a phosphodiester bond. The compound did not showed any hydrogen bond but literature reveals that existence of intercalating forces like van der Waals forces or hydrophobic are potentially significant than hydrogen bonding. The resulting decrease in energy of steroid-DNA complex is –276.5 KJ mol-1 which proves the stable and potent interaction between duplex and steroid molecules.
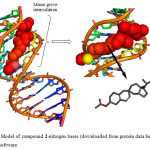 |
Figure4: Model of compound 2-nitrogen bases (downloaded from protein data bank) done by HEX 6.1 software. Click here to View Figure |
Physicochemical properties
Bioactivity score
The bioactivity scores of the compound was also calculated for six parameters, GPCR ligand activity, ion channel modulation, kinase inhibition activity, protease inhibitor, enzyme inhibitor and nuclear receptor ligand activity. As we know for organic molecules, if the bioactivity score is more than 0.00 then, the compound is active, but if it is between -0.50 and 0.00 then the compound is moderately active and if the compound has -0.50 then it is inactive compound. The potential bioactivity score of the compound is given in (Table 2) which clearly show that compound show those properties which are required for the characteristics of compound for acting as a drug.
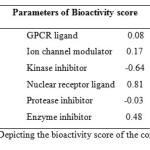 |
Table 2: Depicting the bioactivity score of the compoundClick here to View table |
PASS Analysis
Molecular properties such as membrane permeability and bioavailability are always associated with some basic molecular descriptors such as log P (partition coefficient), molecular weight (MW), hydrogen bond acceptors and donors count in a molecule. Lipinski used these molecular properties in formulating his ‘‘Rule of Five’’. This rule states that most molecules with good membrane permeability have log P ≤ 5, molecular weight ≤ 500, number of hydrogen bond acceptors ≤ 10 and number of hydrogen bond donors ≤ 5. This rule is widely used as a filter for drug-like properties. Although the required molecule did not showed better permeability by showing log P > 5 but other parameters like topological surface area, molecular weight, hydrogen bond acceptor and hydrogen bond donor show that compound has potential to act as drug (Table 3). Also by considering the bioactivity score, the overall potential of the compound can be said to be physiologically active.
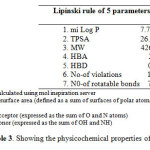 |
Table3: Showing the physicochemical properties of the compound Click here to View table |
Fluorescence spectral studies
At room temperature, compound emit luminescence at 324 nm respectively in 0.01 tris-HCl/50mM NaCl buffer when excited at 260 nm. Fixed amount of compound was titrated with increasing amount of CT DNA, over a range of 5.3 – 2.6 × 10-5 M DNA concentration. The addition of DNA caused a gradual decrease in fluorescence emission (Fig. 5) intensity being consistent with the non-intercalative binding mode such as electrostatic binding mode (surface binding) and be protected by DNA efficiently, since the hydrophobic environment inside the DNA helix reduces the accessibility of solvent molecule to the complex, which as a consequence restrict the complex mobility at binding site, leading to a decrease of the vibrational modes of relaxation. Also, the number of bound steroid molecules per DNA (n) calculated for was found to be 1.06 from fluorescence spectral data. The binding constant K, determined from the Scatchard equation for compound was calculated to be 1.68 × 103 M-1.
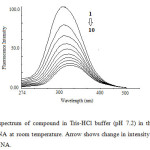 |
Fig 5: Emission spectrum of compound in Tris-HCl buffer (pH 7.2) in the presence and absence of CT DNA at room temperature. Arrow shows change in intensity with increasing concentration of DNA. Click here to View Figure |
EXPERIMENTAL
General
IR spectrum was determined in KBr with Perkins Elmer 237 spectrophotometer, 1H NMR spectrum were run in CDCl3 on Bruker AV 400. 13C NMR was also run in CDCl3 on Bruker DRX 300 and its values are given in ppm. Reaction completion was checked by thin layer chromatography Acetate of the cholecalciferol (vit D3) (commercial substance) was prepared by acetylation of the corresponding sterol with acetic anhydride and pyridine.
Synthesis of 3β-Acetoxy-9, 10-seco-19, 8(8)-spiro-5(10), 6-cholestadiene (2)
To a solution of cholecalciferol acetate (1 mol) in toluene, was added 2, 3-dimethylbutadiene (1 mol) and BF3-etherate / BF3·OEt2/ SnCl4/ ZnBr2/ p-TsOH (2 mol) The refluxing was continued for 2-3 h. After completion, the crude yield was obtained after removing solvent under reduced pressure that was later extracted with ether, recrystallized from ethanol to give an oily compound 2
Characterization
TLC: Rf = 0.71 (Petrol/ether) (3:1); IR (v cm-1) 1026 cm-1 (C-O), 1735 cm-1 (OCOCH3), 1655, 1629 cm-1 for diene system. 1H NMR (500 MHz) δ 5.2 (m, 1H, C3–αH), δ 5.8 (d, 1H, CH), 6.8 (d, 1H, CH), 2.1 (s, 3H, OCOCH3), 0.69 (s, 3H, C13-CH3). 0.96, 0.89 and 0.81 (other CH3); 13C NMR (125 MHz) δ (26.5) C1, (27.7) C2, (71.1) C3, (36.3) C4, (135.1) C5, (138.2) C6, (126.2) C7, (35.5) C8, (45.3) C9, (128.2) C10, (21.5) C11, (38.4) C12, (39.5) C13, (55.5) C14, (25.9) C15, (28.8) C16, (56.5) C17, (14.5) C18, (12.7) C19, (29.6) C20, (18.4) C21, (36.4) C22, (24.2) C23, (38.2) C24, (28.2) C25, (22.5) C26, (23.5) C27, (170.9), (170.9) C1’,(21.5) C2’. MS: m/z 426[M+.].
Evaluation of In Vitro DNA Damage using Plasmid PBR322
The mixture of buffer (pH7.5, 30 µl), 0.5 µg PBR322 DNA and 100 µM compound 2 was prepared. The 20 µl solution containing 40 mM ethylenediaminetetraaectic acid, 0.05% bromophenol blue and 50% (v/v) glycerol was added after incubation for 1 h. The mixture was electrophoresized in submarine 1% agarose paste. UV-transilluminator was used to view and photograph the Ethidium bromide stained gel.
Evaluation of Ex Vivo DNA Damage by Comet Assay using Human Lymphocytes
Comet assay confirmed by the method incarnated and used by Rizvi and co-workers [18] with certain changes. Normal melting agarose was used to precoat the fully frosted slides and to form a cell suspension, few micro litres of agarose (low m.p) and lymphocytes were mixed and put over the former paste and were put hidden. To solidify agarose, the strains were cooled down for few min in a refrigerator. The removal of cover slips was done gently and another coating of 0.5% agarose (low M.p) was drawn over the already solidified layer. For solidification, the covered strains were put on the slides and drawn in cold. The removal of covered slips was done and the ice cold lysis buffer was taken in which slides were immersed in for an hour. After lysis in basic electrophoretic medium, the DNA was allowed to unwind. The gel documentation system was worked out at 4 °C in field strength of 0.7 V/cm and 300 mA current. The strains got neutralized with ice cold 0.4 M buffer pH 7.5, stained with 75 µl EtBr (20 mg/ml) and covered with a cover slip. A fluorescent microscope incorporated with CC camera was used to picture the images of strains. Analysis included images of 25 cells in each triplicate. DNA disintegration was measured in terms of the tail length.
Molecular Docking
HEX 6.1 software was used to proclaim the molecular docking studies [19]. HEX is a software program for analyzing the binding energy, depicting stereo chemical interacting modes of ligand with DNA and stability of docked complex. It studies the interaction between the heterocycle and the host (DNA [d(CGCAAATTTCGC)2 (PDB ID: 1BNA)] /protein) (as input) in protein data bank format and the output is a stable complex deduced with shape complementary and decrease in binding energy criteria. The compound structure was generated by Avagadro 1.01 software using MMFF94 force field. Visualization of the interacted complex has been done by using PyMol (http://pymol.sourceforge.net/) graphic programming [20]. An Intel CORE i5, 2.6 GHz based computer was used to sort the calculations
Determination of physicochemical properties
Bioactivity score and PASS (Prediction of biological activity spectra) analysis
The physicochemical parameters including octanol partition coefficients (miLogP), Mw, HBD, HBA, TPSA and Rotatable bonds were calculated using molinspiration server (http://www.molinspiration.com/cgi-bin/properties) and ChemAxon (chemicalize.org). The PASS (Prediction of biological activity spectra) approach is based on the structure-activity correlation, Compound activity prediction is done by ‘‘comparing’’ the structure of query compound with the structure of well-known biological active substrate existing in database of the freely available PASS web service. Molecule activity prediction is done by ‘‘comparing’’ the structure of query compound with the structure of well-known biological active substrate existing in database of PASS web service.
Fluorescence Spectral study
The fluorescence measurements were carried out on a Shimazdu spectrofluorimeter (Model USA), while DNA (5.3 – 2.6 × 10-5) pre-treated with compound (4 × 10-5M) for 30 min in buffer. In the fixed concentration of compound, the emission intensities of DNA were recorded between 274 and 500 nm, at the excitation wavelength of 260 nm.
CONCLUSION
A convenient and facile route for synthesizing new cholestadiene derivative has been reported. The gel electrophoresis depicted that compound (in comparison to 5-fluorouracil) acts to be active DNA cleaving agent. Comet assay depicted that compound exhibited potential cell apoptosis hence proved to be genotoxic nature. The interactive docking studies undertaken revealed minor groove binding of compound to the DNA molecule. The bioactivity score and PASS analysis also predicted the potential of the compound to act as physiologically active drug. The fluorescence studies also agreed the hypochromism behavior of the compound depicting the decrease in the intensity which also correlates the results with the DNA cleaving ability of the compound as shown in gel electrophoresis.
ACKNOWLEDGEMENT
Sultanat, Ayaz M Dar, A. Rizvi and I Naseem are thankful to Chairman of Chemistry Department of AMU, Aligarh for providing research facilities. Dr. Sultanat is thankful to the Department of Science and Technology (DST) New Delhi (India) for providing financial assistance under the women scientist scheme-A.
REFERENCES
- Kovacs, D. Kadar, Z. Motyan, G. Schneider, G. Wolfling, J. Zupko, I. Frank, E. Steroids, 2012, 77, 1075-1085.
- Ballistreri, F. P. Chillemi, R. Sciuto, S. Tomaselli, G. A. Toscano, R. M. Steroids, 2006, 71, 565-577.
- Xin, Y. Jiang, H. Zhao, J. Zhu, S. Tetrahedron, 2008, 64, 9315-9319.
- Jiang, H. L. Zhu, S. Z. Synlett, 2006, 9, 1343-1346.
- Marshall, J. A. Gill, K. J. Organomet. Chem., 2001,624, 294-299.
- Van Leeuwen, S. H. Quaedflieg, P. J. L. M., Broxterman, Q. B. Milhajlovic, Y. Liskamp R. M. J. Tetrahedron Lett., 2005, 46, 653-656.
- Terentev, A. O. Kutkin, A. V. Platonov, M. M. Ogibin, Y. N. Nikishin,G. I. Tetrahedron Lett., 2003,44,7359-7363.
- Kuhnert, N. Clifford, M. N. Radenac, A.G. Tetrahedron Lett., 2001, 42, 9261- 9263.
- Stephan, E. Olaru, A. Jaouen, G. Tetrahedron Lett., 1999, 40, 8571-8574.
- Hardouin, C. Taran, F. Doris, E. J. Org. Chem., 2001,66, 4450- 4452.
- Srikrishna, A. Ramasastry, S. S. V. Tetrahedron Asym., 2005, 16, 2973-2979.
- Chang, M.Y. Lin, C.H. Chen, Y.L. Chang, C.Y. Hsu, R.T. Org.Lett., 2010,12, 1176-1179.
- Nakamura, M. Suzuki, A. Nakatani, M. Fuchikami, T. Inoue, M. Katoh,T. Tetrahedron Lett.; 2002, 43, 6929-6932.
- Gelman, D. M. Forsyth, C. M. Perlmutter, P. Org. Lett., 2009, 11, 4958-4960.
- Yates, P. Eaton P. J. Am. Chem. Soc., 1960, 82, 4436-4437.
- Mushfiq, M. Khan, A.R., Shamim, A. Rehman, R. Sultanat, Syn. Comm., 2010, 40, 1508-1515.
- Nicolaou, K. C. Snyder, Scott, A. Montagnon, Tamsyn, Vassilikogiannakis, Georgios, . Ang. Chem. Int. Ed., 2002, 41. 1668.
- Rizvi, A. Hasan, S. S. Naseem, I. Selective cytotoxic action and DNA Damage by Calcitriol-Cu (II) interaction: Putative mechanism of cancer prevention. PLoS One, 2013, DOI 10.1371/ journal.pone.0076191.
- Mustard, D. Ritchie, D. W. Struct. Funct. Bioinf., 2005, 60, 269-274.
- DeLano, W. L. The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA, USA, 2002.

This work is licensed under a Creative Commons Attribution 4.0 International License.









